
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,332,504 – 22,332,613 |
| Length | 109 |
| Max. P | 0.881300 |

| Location | 22,332,504 – 22,332,613 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -25.11 |
| Energy contribution | -25.83 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22332504 109 + 27905053 AAAAUUCAAAGAGUGAUUUGCCAAGUGCA---CUCGCUUUUGUGAAACCUUUCAAACUUAAUUGCCACAUUAUGUGGCAGUGUUCCAAGUUGAUGCGUCCAACUUGGCAUAU ....((((((((((((..(((.....)))---.)))))))).))))..............((((((((.....))))))))((.((((((((.......)))))))).)).. ( -33.00) >DroSec_CAF1 49263 109 + 1 AAAAUUCAAUGAAUGAUUUGCCAAGUGCA---CUCGCUUUUGUGAAACCUUUCAAACUUAAUUGCCACAUUAUGUGGCAGUGUUCCAAGUUGAUGCGUCCAACUUGGCAUAU ..................((((((((...---..(((.....((((....))))(((((.((((((((.....)))))))).....)))))...)))....))))))))... ( -31.20) >DroSim_CAF1 48274 109 + 1 AAAAUUCAAUGAAUGAUUUGCCAAGUGCA---CUCGCUUUUGUGAAACCUUUCAAACUUAAUUGCCACAUUAUGUGGCAGUGUUCCAAGUUGAUGCGUCCAACUUGGCAUAU ..................((((((((...---..(((.....((((....))))(((((.((((((((.....)))))))).....)))))...)))....))))))))... ( -31.20) >DroEre_CAF1 51286 109 + 1 AAAAAGCUGAGUAUGUUUUGCCAAGUGCA---CUCGCUUUUGUGAAACCUUUCAAACUUAAUUGCCACAUUAUGUGGCAGUGUUCCAAGUUGAUGCGUCCAACUUGGCAUAU ..(((((.......)))))(((((((...---..(((.....((((....))))(((((.((((((((.....)))))))).....)))))...)))....))))))).... ( -32.00) >DroYak_CAF1 50576 109 + 1 AAAAUUCUGAGAAUGUUUUGCCAAGUGCA---CUCGCUAUUGUGAAACCUUUCAAACGUAAUUGCCACAUUAUGUGGCAGUGUUCCAAGUUGAUGCGUCCAACUUGGCAUAU .......(((((..((((..(..((((..---..))))...)..)))).)))))......((((((((.....))))))))((.((((((((.......)))))))).)).. ( -31.30) >DroAna_CAF1 52581 107 + 1 UUCAUUCAA-----AUUUUCACAAGUGCAGCUCUGAUGCUGGGGAAACCUAUUCAACUUAAUUGCCACAUUAUGUGGCUGUGUUCCAAGUUGAUGCGUUCAACUUGGCAUAU .........-----.....(((((((.((((......))))((....))......))))....(((((.....))))).)))..(((((((((.....)))))))))..... ( -34.10) >consensus AAAAUUCAAAGAAUGAUUUGCCAAGUGCA___CUCGCUUUUGUGAAACCUUUCAAACUUAAUUGCCACAUUAUGUGGCAGUGUUCCAAGUUGAUGCGUCCAACUUGGCAUAU ..................(((((((((((....((((....))))......((.(((((.((((((((.....)))))))).....)))))))))).....))))))))... (-25.11 = -25.83 + 0.72)

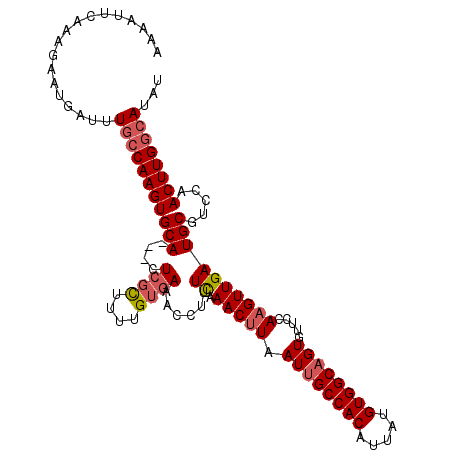
| Location | 22,332,504 – 22,332,613 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -22.58 |
| Energy contribution | -23.17 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22332504 109 - 27905053 AUAUGCCAAGUUGGACGCAUCAACUUGGAACACUGCCACAUAAUGUGGCAAUUAAGUUUGAAAGGUUUCACAAAAGCGAG---UGCACUUGGCAAAUCACUCUUUGAAUUUU ...((((((((.(.((.(.(((((((((.....((((((.....)))))).))))).))))...((((.....))))).)---).))))))))).................. ( -28.80) >DroSec_CAF1 49263 109 - 1 AUAUGCCAAGUUGGACGCAUCAACUUGGAACACUGCCACAUAAUGUGGCAAUUAAGUUUGAAAGGUUUCACAAAAGCGAG---UGCACUUGGCAAAUCAUUCAUUGAAUUUU ...((((((((.(.((.(.(((((((((.....((((((.....)))))).))))).))))...((((.....))))).)---).)))))))))..(((.....)))..... ( -29.40) >DroSim_CAF1 48274 109 - 1 AUAUGCCAAGUUGGACGCAUCAACUUGGAACACUGCCACAUAAUGUGGCAAUUAAGUUUGAAAGGUUUCACAAAAGCGAG---UGCACUUGGCAAAUCAUUCAUUGAAUUUU ...((((((((.(.((.(.(((((((((.....((((((.....)))))).))))).))))...((((.....))))).)---).)))))))))..(((.....)))..... ( -29.40) >DroEre_CAF1 51286 109 - 1 AUAUGCCAAGUUGGACGCAUCAACUUGGAACACUGCCACAUAAUGUGGCAAUUAAGUUUGAAAGGUUUCACAAAAGCGAG---UGCACUUGGCAAAACAUACUCAGCUUUUU .....(((((((((.....))))))))).....((((((.....))))))........((((....)))).(((((((((---((....((......))))))).)))))). ( -29.50) >DroYak_CAF1 50576 109 - 1 AUAUGCCAAGUUGGACGCAUCAACUUGGAACACUGCCACAUAAUGUGGCAAUUACGUUUGAAAGGUUUCACAAUAGCGAG---UGCACUUGGCAAAACAUUCUCAGAAUUUU ...((((((((.(.((.........((((((..((((((.....))))))....(........))))))).........)---).))))))))).................. ( -28.67) >DroAna_CAF1 52581 107 - 1 AUAUGCCAAGUUGAACGCAUCAACUUGGAACACAGCCACAUAAUGUGGCAAUUAAGUUGAAUAGGUUUCCCCAGCAUCAGAGCUGCACUUGUGAAAAU-----UUGAAUGAA .....(((((((((.....)))))))))......(((((.....)))))..(((((((..((((((.....((((......)))).))))))...)))-----))))..... ( -32.60) >consensus AUAUGCCAAGUUGGACGCAUCAACUUGGAACACUGCCACAUAAUGUGGCAAUUAAGUUUGAAAGGUUUCACAAAAGCGAG___UGCACUUGGCAAAACAUUCUUUGAAUUUU ...((((((((.....(((....((((......((((((.....))))))........((((....))))......))))...))))))))))).................. (-22.58 = -23.17 + 0.58)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:56 2006