
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,325,174 – 22,325,275 |
| Length | 101 |
| Max. P | 0.828191 |

| Location | 22,325,174 – 22,325,275 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.94 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -21.75 |
| Energy contribution | -23.12 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
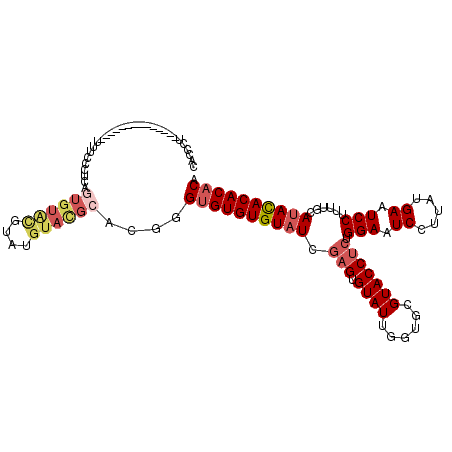
>3R_DroMel_CAF1 22325174 101 + 27905053 CACCCU--------------UUUCCCUCAGUGUACGUAUGUACGCACGGGUGUGUGUAUUGAGUGUAUUGGUGCGUACCUCUGGAAUCCUUAUGAAUCCU-UUGCAUAUACACACA ......--------------...(((...((((((....))))))..)))(((((((((...(((((..((..((((.....((...)).))))...)).-.)))))))))))))) ( -28.30) >DroSec_CAF1 42020 102 + 1 CACCCU--------------UUUCCCUCAGUGUACGUAUGUACGCACGGGUGUGUGUAUUGAGUGUAUUGGUGCGUACCUCUGGAAUCCUUAUGAAUCCUUUUGCAUACACACACA ......--------------....((...((((((....))))))..))((((((((((.(((.((((......))))))).(((.((.....)).)))......)))))))))). ( -29.80) >DroSim_CAF1 40979 102 + 1 CACCCU--------------UUUCCCUCAGUGUACGCAUGUACGCACGGGUGUGUGUAUUGAGUGUAUUGGUGCGUACCUCUGGAAUCCUUAUGAAUCCUUUUGCAUACACACACA ......--------------.........(((((.(((.((((((((.((((..(.......)..)))).))))))))....(((.((.....)).)))...))).)))))..... ( -30.10) >DroEre_CAF1 43372 102 + 1 ACCCUU--------------UUUCCCUCAGUGUGCGAGUGUACGCACGGGUGUGUGUAUCGAGUGUAUUGGUGCGUACCUCUGGAAUCCUUAUGAAUCCUUUUGCAUACACACACA ......--------------.........(((((((((.((((((((.((((..(.......)..)))).))))))))))).(((.((.....)).)))....))))))....... ( -34.10) >DroYak_CAF1 42940 102 + 1 ACUCCU--------------UUUCUCUCAAUGUAUGGAUGUAUGCACGGGUGUGUGUAUCGAGUGUAUUGGUGCGUACCUCUGGAAUCCUUAUGAAUCCUUUUGCAUACACACACA ......--------------..............((..((((((((.(((.(((((((((((.....)))))))))))))).(((.((.....)).)))...))))))))..)).. ( -27.70) >DroAna_CAF1 43412 103 + 1 UUCCUUGCCGAGAAACAGAAUUUUCCUUAGUGUCU-------------UGAGUGUGUAUCAAGUGUAUUGGUGCGUACCCCUGGAAUCCUUAUGAAUCCUUUCGCACACACACACA ........(((((.((.((.......)).)).)))-------------)).((((((..(((.....)))(((((.......(((.((.....)).)))...)))))))))))... ( -22.70) >consensus CACCCU______________UUUCCCUCAGUGUACGUAUGUACGCACGGGUGUGUGUAUCGAGUGUAUUGGUGCGUACCUCUGGAAUCCUUAUGAAUCCUUUUGCAUACACACACA .............................((((((....))))))....((((((((((.(((.((((......))))))).(((.((.....)).)))......)))))))))). (-21.75 = -23.12 + 1.36)


| Location | 22,325,174 – 22,325,275 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.94 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -23.77 |
| Energy contribution | -24.62 |
| Covariance contribution | 0.85 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
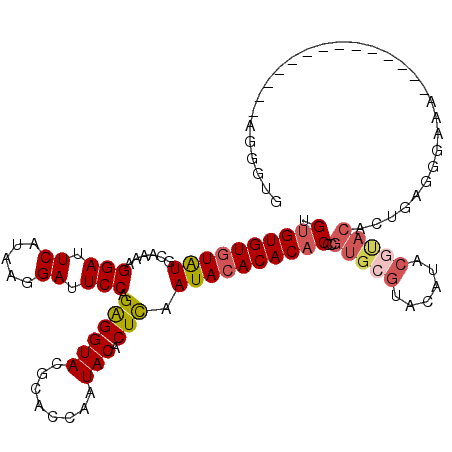
>3R_DroMel_CAF1 22325174 101 - 27905053 UGUGUGUAUAUGCAA-AGGAUUCAUAAGGAUUCCAGAGGUACGCACCAAUACACUCAAUACACACACCCGUGCGUACAUACGUACACUGAGGGAAA--------------AGGGUG .((((((((......-.(((.((.....)).))).((((((........))).))).))))))))((((((((((....)))))).(....)....--------------.)))). ( -31.10) >DroSec_CAF1 42020 102 - 1 UGUGUGUGUAUGCAAAAGGAUUCAUAAGGAUUCCAGAGGUACGCACCAAUACACUCAAUACACACACCCGUGCGUACAUACGUACACUGAGGGAAA--------------AGGGUG ...((((((((......(((.((.....)).))).((((((........))).))).))))))))((((((((((....)))))).(....)....--------------.)))). ( -31.10) >DroSim_CAF1 40979 102 - 1 UGUGUGUGUAUGCAAAAGGAUUCAUAAGGAUUCCAGAGGUACGCACCAAUACACUCAAUACACACACCCGUGCGUACAUGCGUACACUGAGGGAAA--------------AGGGUG ...((((((((......(((.((.....)).))).((((((........))).))).))))))))((((((((((....)))))).(....)....--------------.)))). ( -31.40) >DroEre_CAF1 43372 102 - 1 UGUGUGUGUAUGCAAAAGGAUUCAUAAGGAUUCCAGAGGUACGCACCAAUACACUCGAUACACACACCCGUGCGUACACUCGCACACUGAGGGAAA--------------AAGGGU ...((((((((.(....(((.((.....)).))).((((((........))).))))))))))))(((((((((......))))).(....)....--------------..)))) ( -31.50) >DroYak_CAF1 42940 102 - 1 UGUGUGUGUAUGCAAAAGGAUUCAUAAGGAUUCCAGAGGUACGCACCAAUACACUCGAUACACACACCCGUGCAUACAUCCAUACAUUGAGAGAAA--------------AGGAGU ((((((((((((((...(((.((.....)).))).((((((........))).)))..............))))))))..))))))..........--------------...... ( -24.30) >DroAna_CAF1 43412 103 - 1 UGUGUGUGUGUGCGAAAGGAUUCAUAAGGAUUCCAGGGGUACGCACCAAUACACUUGAUACACACUCA-------------AGACACUAAGGAAAAUUCUGUUUCUCGGCAAGGAA .((((((((((((....(((.((.....)).)))....)))))))).......(((((.......)))-------------)))))).........((((((......)).)))). ( -23.80) >consensus UGUGUGUGUAUGCAAAAGGAUUCAUAAGGAUUCCAGAGGUACGCACCAAUACACUCAAUACACACACCCGUGCGUACAUACGUACACUGAGGGAAA______________AGGGUG .((((((((((......(((.((.....)).))).((((((........))).))).))))))))))..(((((......)))))............................... (-23.77 = -24.62 + 0.85)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:52 2006