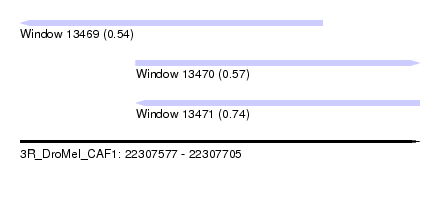
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,307,577 – 22,307,705 |
| Length | 128 |
| Max. P | 0.739783 |

| Location | 22,307,577 – 22,307,674 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.33 |
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -13.89 |
| Energy contribution | -15.43 |
| Covariance contribution | 1.54 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22307577 97 - 27905053 GAUUCAAGUU----------GCU-GCUGCAGUUGUUGCAGUUGUGCCGUUGCUGGUGCAGCAGCAACACGAGCUACAAGAUACUUUCUGUGCAACCAUAGAACUUCCC .......(((----------(((-((((((.(.((.((.(.....).)).)).).)))))))))))).................(((((((....)))))))...... ( -31.80) >DroSec_CAF1 24001 97 - 1 GAUUCAAGUU----------GCU-GCUGCAGUUGUUGCAGUUGUGCAGUUGCUGGUGCAGCAGCAACACGAGCUACAAGAUACUUUCUGUGCAACCAUAGAACUUCCC .......(((----------(((-((((((((..(((((....)))))..))...)))))))))))).................(((((((....)))))))...... ( -33.30) >DroSim_CAF1 23489 97 - 1 GAUUCAAGUU----------GCU-GCUGCAGUUGUUGCAGUUGUGCAGUUGCUGGUGCAGCAGCAACACGAGCUACAAGAUACUUUCUGUGCAACCAUAGAACUUCCC .......(((----------(((-((((((((..(((((....)))))..))...)))))))))))).................(((((((....)))))))...... ( -33.30) >DroEre_CAF1 25027 97 - 1 GAUUCAAGUU----------GCU-GCUGCAGUUGUUGCAGUUGUGCAGUUGCUGUUGCAGCAGCAACACGAGCUACAAGAUACUUUCUGUGCAACCAUAGAACUUCCC .......(((----------(((-(((((((..((((((....))))...))..))))))))))))).................(((((((....)))))))...... ( -33.50) >DroWil_CAF1 26738 107 - 1 GCUUGUUGUUGUUUGUUGUUGCU-GUAGCAAUUGUUAGAGUUGAACGGCAGCAGCAGCAGCAGCAACUUGAGCUACAAGAUACUUUCUGUGCAACCGUAGAACUUCCC (((((((((((((.(((((((((-((..(((((.....)))))....))))))))))))))))))))..))))...........(((((((....)))))))...... ( -37.40) >DroAna_CAF1 23808 81 - 1 GAUUCAAGUU----------GCAUAGCGAUGAUGUUGCUGUU--------------GU---UGCAACUUCAGCUACAAGAUACUUUCUGUGCAACCAUAGAACUUCCC .......(((----------(((((((..(((.(((((....--------------..---.))))).))))))...(((.....))))))))))............. ( -20.10) >consensus GAUUCAAGUU__________GCU_GCUGCAGUUGUUGCAGUUGUGCAGUUGCUGGUGCAGCAGCAACACGAGCUACAAGAUACUUUCUGUGCAACCAUAGAACUUCCC ....................((.....))..((((.((.(((((...(((((....))))).)))))....)).))))......(((((((....)))))))...... (-13.89 = -15.43 + 1.54)


| Location | 22,307,614 – 22,307,705 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 78.03 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -8.98 |
| Energy contribution | -11.17 |
| Covariance contribution | 2.18 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22307614 91 + 27905053 UCGUGUUGCUGCUGCACCAGCAACGGCACAACUGCAACAACUGCAGC-AGC----------AACUUGAAUCGCAUCUAUUGAUUGCACACAGUUCAUUGGCG (((.((((((((((((.........(((....)))......))))))-)))----------))).)))..(((....((.(((((....))))).))..))) ( -29.26) >DroSec_CAF1 24038 91 + 1 UCGUGUUGCUGCUGCACCAGCAACUGCACAACUGCAACAACUGCAGC-AGC----------AACUUGAAUCGCAUCUAUUGAUUGCACACAGUUCAUUGGCG (((.((((((((((((........((((....)))).....))))))-)))----------))).)))..(((....((.(((((....))))).))..))) ( -29.72) >DroSim_CAF1 23526 91 + 1 UCGUGUUGCUGCUGCACCAGCAACUGCACAACUGCAACAACUGCAGC-AGC----------AACUUGAAUCGCAUCUAUUGAUUGCACACAGUUCAUUGGCG (((.((((((((((((........((((....)))).....))))))-)))----------))).)))..(((....((.(((((....))))).))..))) ( -29.72) >DroEre_CAF1 25064 91 + 1 UCGUGUUGCUGCUGCAACAGCAACUGCACAACUGCAACAACUGCAGC-AGC----------AACUUGAAUCGCAUCUAUUGAUUGCACACAGUUCAUUGGCG (((.((((((((((((........((((....)))).....))))))-)))----------))).)))..(((....((.(((((....))))).))..))) ( -29.72) >DroWil_CAF1 26775 101 + 1 UCAAGUUGCUGCUGCUGCUGCUGCCGUUCAACUCUAACAAUUGCUAC-AGCAACAACAAACAACAACAAGCACAUCUAUUGAUUGCAUGCAGUUCAUUGGCG ((((...(((((((.(((((..((.(((..........))).))..)-)))).))..............(((.(((....))))))..)))))...)))).. ( -23.00) >DroAna_CAF1 23845 75 + 1 UGAAGUUGCA---AC--------------AACAGCAACAUCAUCGCUAUGC----------AACUUGAAUCGCAUCCAUUGAUUGCACACAGUUCAUUGGCG (((.(((((.---..--------------....))))).))).(((((((.----------(((((((((((.......))))).))...)))))).))))) ( -16.50) >consensus UCGUGUUGCUGCUGCACCAGCAACUGCACAACUGCAACAACUGCAGC_AGC__________AACUUGAAUCGCAUCUAUUGAUUGCACACAGUUCAUUGGCG ....(((((((......))))))).......(((((.....))))).............................(((.((((((....))).))).))).. ( -8.98 = -11.17 + 2.18)


| Location | 22,307,614 – 22,307,705 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 78.03 |
| Mean single sequence MFE | -31.05 |
| Consensus MFE | -12.55 |
| Energy contribution | -14.12 |
| Covariance contribution | 1.57 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22307614 91 - 27905053 CGCCAAUGAACUGUGUGCAAUCAAUAGAUGCGAUUCAAGUU----------GCU-GCUGCAGUUGUUGCAGUUGUGCCGUUGCUGGUGCAGCAGCAACACGA ........(((((((.((((((....)))(((((....)))----------)))-)))))))))(((((.((((..(((....)))..)))).))))).... ( -32.00) >DroSec_CAF1 24038 91 - 1 CGCCAAUGAACUGUGUGCAAUCAAUAGAUGCGAUUCAAGUU----------GCU-GCUGCAGUUGUUGCAGUUGUGCAGUUGCUGGUGCAGCAGCAACACGA ......((((.....((((.((....)))))).)))).(((----------(((-((((((((..(((((....)))))..))...)))))))))))).... ( -32.30) >DroSim_CAF1 23526 91 - 1 CGCCAAUGAACUGUGUGCAAUCAAUAGAUGCGAUUCAAGUU----------GCU-GCUGCAGUUGUUGCAGUUGUGCAGUUGCUGGUGCAGCAGCAACACGA ......((((.....((((.((....)))))).)))).(((----------(((-((((((((..(((((....)))))..))...)))))))))))).... ( -32.30) >DroEre_CAF1 25064 91 - 1 CGCCAAUGAACUGUGUGCAAUCAAUAGAUGCGAUUCAAGUU----------GCU-GCUGCAGUUGUUGCAGUUGUGCAGUUGCUGUUGCAGCAGCAACACGA ...........((((((((.((....))))).......(((----------(((-((.(((((..(((((....)))))..))))).)))))))).))))). ( -34.70) >DroWil_CAF1 26775 101 - 1 CGCCAAUGAACUGCAUGCAAUCAAUAGAUGUGCUUGUUGUUGUUUGUUGUUGCU-GUAGCAAUUGUUAGAGUUGAACGGCAGCAGCAGCAGCAGCAACUUGA ............((((...(((....)))))))(((((((((((.(((((((((-((..(((((.....))))).))))))))))))))))))))))..... ( -35.40) >DroAna_CAF1 23845 75 - 1 CGCCAAUGAACUGUGUGCAAUCAAUGGAUGCGAUUCAAGUU----------GCAUAGCGAUGAUGUUGCUGUU--------------GU---UGCAACUUCA ......((((.(((((.((.....)).))))).))))((((----------((((((((((...)))))))..--------------..---)))))))... ( -19.60) >consensus CGCCAAUGAACUGUGUGCAAUCAAUAGAUGCGAUUCAAGUU__________GCU_GCUGCAGUUGUUGCAGUUGUGCAGUUGCUGGUGCAGCAGCAACACGA .((((((.((((...((((.((....)))))).....))))..........((.....)).))))..)).(((((...(((((....))))).))))).... (-12.55 = -14.12 + 1.57)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:39 2006