
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,292,978 – 22,293,085 |
| Length | 107 |
| Max. P | 0.981374 |

| Location | 22,292,978 – 22,293,085 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.61 |
| Mean single sequence MFE | -37.47 |
| Consensus MFE | -18.77 |
| Energy contribution | -18.92 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
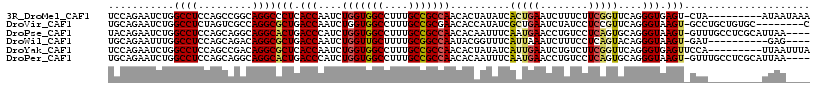
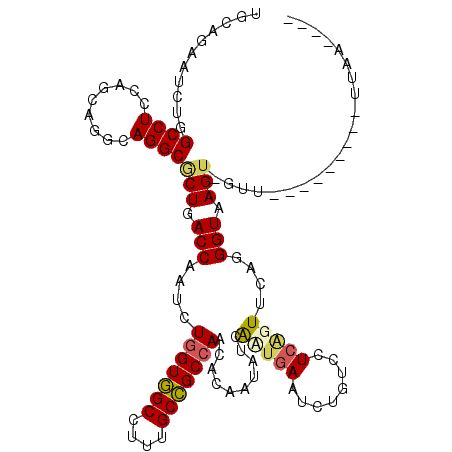
>3R_DroMel_CAF1 22292978 107 + 27905053 UCCAGAAUCUGGCCUCCAGCCGGCAGGCCCUCACCAAUCUGGUGGCCUUUGCCGCCAACACUAUAUCACUGAAUCUUUCUUCGGUUCAGGGUGAGU-CUA---------AUAAUAAA ...(((....(((((((....)).)))))((((((....(((((((....)))))))...........(((((((.......))))))))))))))-)).---------........ ( -41.00) >DroVir_CAF1 10970 108 + 1 UGCAGAAUCUGGCCUCUAGUCGCCAGGCGCUGACCAAUCUGGUGGCCUUUGCCGCGAACACCAUAUCGCUGAAUCUAUCCUCCGUUCAGGGUAAGU-GCCUGCUGUGC--------C .((((...(((((........)))))(((((.(((......(((((....))))).............((((((.........))))))))).)))-)))))).....--------. ( -35.90) >DroPse_CAF1 9092 112 + 1 UACAGAAUCUGGCCUCCAGCAGGCAGGCACUGACCCAUCUGGUGGCCUUUGCCGCCAACACAAUUUCAAUGAACCUGUCCUCAGUGCAGGGUAAGU-GUUUGCCUCGCAUUAA---- ..(((...))).......(((((((((((((.((((...(((((((....)))))))............((...(((....)))..)))))).)))-)))))))).)).....---- ( -42.40) >DroWil_CAF1 10768 102 + 1 UGCAGAAUUUGGCCUCCAGCAGACAGGCGCUGACCAAUCUGGUUGCUUUUGCGGCCAAUACGGUUUCAUUAAAUCUUUCCUCAGUACAGGGUAAGU-GAU----------GAG---- ..((..(((((.(((...((......))(((((......(((((((....)))))))....(((((....))))).....)))))..))).)))))-..)----------)..---- ( -25.50) >DroYak_CAF1 8911 108 + 1 UCCAGAAUCUGGCCUCCAGCCGACAGGCGCUCACCAAUCUGGUGGCCUUUGCCGCCAACACUAUAUCAUUGAAUCUGUCUUCGGUUCAGGGUGAGUUCCA---------UUAAUUUA .........((((.....))))...((.(((((((....(((((((....)))))))...........(((((((.......)))))))))))))).)).---------........ ( -36.30) >DroPer_CAF1 9220 112 + 1 UGCAGAAUCUGGCCUCCAGCAGGCAGGCACUGACCCAUCUGGUGGCCUUUGCCGCCAACACAAUUUCAAUGAACCUGUCCUCAGUGCAGGGUAAGU-GUUUGCCUCGCAUUAA---- (((.....((((...)))).(((((((((((.((((...(((((((....)))))))............((...(((....)))..)))))).)))-)))))))).)))....---- ( -43.70) >consensus UGCAGAAUCUGGCCUCCAGCAGGCAGGCGCUGACCAAUCUGGUGGCCUUUGCCGCCAACACAAUAUCAAUGAAUCUGUCCUCAGUUCAGGGUAAGU_GUU_________UUAA____ ...........((((.........))))(((.(((....(((((((....)))))))..........(((((........)))))....))).)))..................... (-18.77 = -18.92 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:30 2006