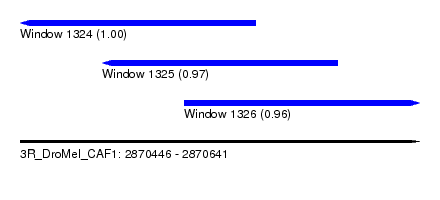
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,870,446 – 2,870,641 |
| Length | 195 |
| Max. P | 0.996456 |

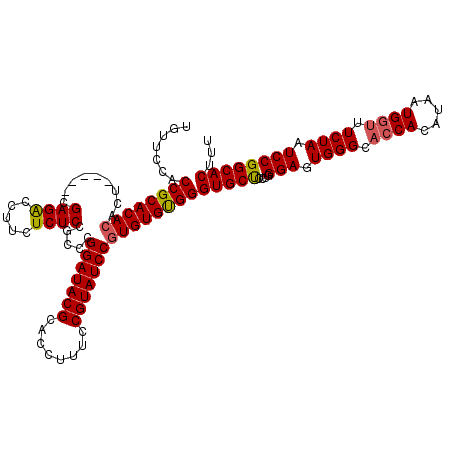
| Location | 2,870,446 – 2,870,561 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.59 |
| Mean single sequence MFE | -41.77 |
| Consensus MFE | -40.39 |
| Energy contribution | -40.57 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.70 |
| SVM RNA-class probability | 0.996456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2870446 115 - 27905053 UGUUCCACCGCACACACU-----CGAGACCUUCUCUCCGCCCGGAUACGCACCUUUCCGUAUCCGUGUGUGGGUGCUCCUGGGAGUGGGCACCACAUAAUGGUUUCUAAUCCGGCACUUU .......((((((((...-----.((((.....)))).....(((((((........)))))))))))))))(((((....(((.((((.((((.....)))).)))).))))))))... ( -43.30) >DroSec_CAF1 1418 120 - 1 UGUUCCACCGCACACACUCGACUCGAGACCUUCUCUCCGCCCGGAUACGCACCUUUCCGUAUCCGUGUGUGGGUGCUCCUGGGAGUGGGCACCACAUAAUGGCUUCUAAUCCGGCACUUU .......((((((((.((((...))))...............(((((((........)))))))))))))))(((((....(((.((((..(((.....)))..)))).))))))))... ( -41.40) >DroSim_CAF1 1787 120 - 1 UGUUCCACCGCACACACUCGACUCGAGACCUUCUCUCCGCCCGGAUACGCACCUUUCCGUAUCCGUGUGCGGGUGCUCCAGGGAGUGGGCACCACAUAAUGGUUUCUAAUCCGGCACUUU .......((((((((.((((...))))...............(((((((........)))))))))))))))(((((....(((.((((.((((.....)))).)))).))))))))... ( -45.40) >DroEre_CAF1 2340 115 - 1 UGUUCCACCGCACACACU-----CGAGGCCUUCUCUCCGCCUGGAUACGCACCUUUCCGUAUCCCUGUGUGGGUGCCUCUGGGAAUGGGUAGCACAUAAUGGUUUCUAAUCCGGCACUUU .......(((((((....-----..((((.........))))(((((((........))))))).)))))))(((((....(((.((((.(((........))))))).))))))))... ( -37.00) >consensus UGUUCCACCGCACACACU_____CGAGACCUUCUCUCCGCCCGGAUACGCACCUUUCCGUAUCCGUGUGUGGGUGCUCCUGGGAGUGGGCACCACAUAAUGGUUUCUAAUCCGGCACUUU .......((((((((.........((((.....)))).....(((((((........)))))))))))))))(((((....(((.((((.((((.....)))).)))).))))))))... (-40.39 = -40.57 + 0.19)

| Location | 2,870,486 – 2,870,601 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.27 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -35.56 |
| Energy contribution | -35.50 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2870486 115 - 27905053 CCAAGGAUGAAUGAGGAUACCCGAACAUGACACUUAUGGAUGUUCCACCGCACACACU-----CGAGACCUUCUCUCCGCCCGGAUACGCACCUUUCCGUAUCCGUGUGUGGGUGCUCCU ...((((.......((....))((((((..((....)).))))))((((.(((((((.-----.((((.....)))).....(((((((........)))))))))))))))))).)))) ( -38.70) >DroSec_CAF1 1458 120 - 1 CCAAGGAUGAAUGAGGAUACCCGAACAUGACACUUAUGGAUGUUCCACCGCACACACUCGACUCGAGACCUUCUCUCCGCCCGGAUACGCACCUUUCCGUAUCCGUGUGUGGGUGCUCCU ...((((.......((....))((((((..((....)).))))))((((.(((((((.......((((.....)))).....(((((((........)))))))))))))))))).)))) ( -38.70) >DroSim_CAF1 1827 120 - 1 CCAAGGAUGAAUGAGGAUACCCGAACAUGACACUUAUGGAUGUUCCACCGCACACACUCGACUCGAGACCUUCUCUCCGCCCGGAUACGCACCUUUCCGUAUCCGUGUGCGGGUGCUCCA ..............((((((((((((((..((....)).))))))....((((((.((((...))))...............(((((((........)))))))))))))))))).))). ( -41.20) >DroEre_CAF1 2380 115 - 1 CCAAGGAUGAAUGAGGAUACCCGAACAUGACACUUAUGGAUGUUCCACCGCACACACU-----CGAGGCCUUCUCUCCGCCUGGAUACGCACCUUUCCGUAUCCCUGUGUGGGUGCCUCU ............((((......((((((..((....)).))))))((((.((((((..-----..((((.........))))(((((((........))))))).)))))))))))))). ( -37.00) >consensus CCAAGGAUGAAUGAGGAUACCCGAACAUGACACUUAUGGAUGUUCCACCGCACACACU_____CGAGACCUUCUCUCCGCCCGGAUACGCACCUUUCCGUAUCCGUGUGUGGGUGCUCCU ..............((((((((((((((..((....)).))))))....((((((.........((((.....)))).....(((((((........)))))))))))))))))).))). (-35.56 = -35.50 + -0.06)

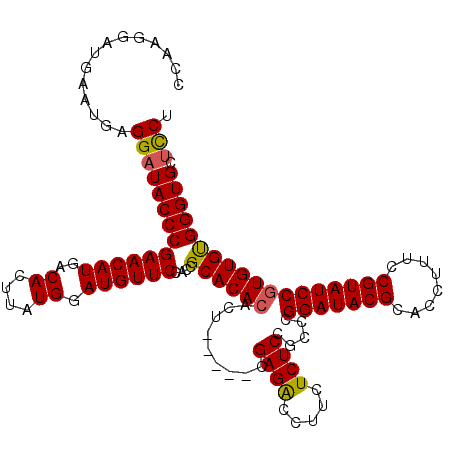
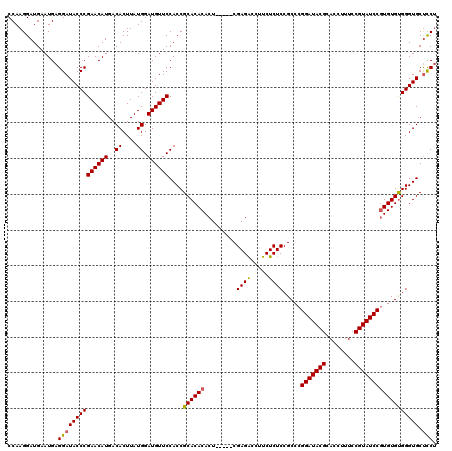
| Location | 2,870,526 – 2,870,641 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.36 |
| Mean single sequence MFE | -43.55 |
| Consensus MFE | -39.82 |
| Energy contribution | -41.82 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
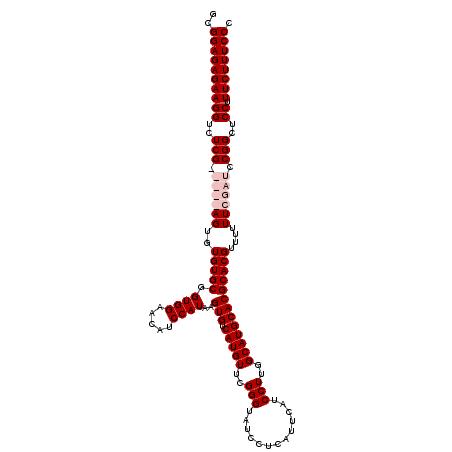
>3R_DroMel_CAF1 2870526 115 + 27905053 GCGGAGAGAAGGUCUCG-----AGUGUGUGCGGUGGAACAUCCAUAAGUGUCAUGUUCGGGUAUCCUCAUUCAUCCUUGGCAUGCACGCACGUUUUUUCGAUCCGGUUCCUUUCUUUCCC ..((((((((((..(((-----((..(((((.((((.....))))..(((.(((((..(((.............)))..)))))))))))))....))))).))(....).)))))))). ( -38.72) >DroSec_CAF1 1498 120 + 1 GCGGAGAGAAGGUCUCGAGUCGAGUGUGUGCGGUGGAACAUCCAUAAGUGUCAUGUUCGGGUAUCCUCAUUCAUCCUUGGCAUGCACGCACGUUUUUUCGAUCCGGCUCCUUUCUUUCCC ..((((((((((..(((..(((((..(((((.((((.....))))..(((.(((((..(((.............)))..)))))))))))))....)))))..)))..)).)))))))). ( -46.62) >DroSim_CAF1 1867 120 + 1 GCGGAGAGAAGGUCUCGAGUCGAGUGUGUGCGGUGGAACAUCCAUAAGUGUCAUGUUCGGGUAUCCUCAUUCAUCCUUGGCAUGCACGCACGUUUUUUCGAUCCGGCUCCUUUCUUUCCC ..((((((((((..(((..(((((..(((((.((((.....))))..(((.(((((..(((.............)))..)))))))))))))....)))))..)))..)).)))))))). ( -46.62) >DroEre_CAF1 2420 115 + 1 GCGGAGAGAAGGCCUCG-----AGUGUGUGCGGUGGAACAUCCAUAAGUGUCAUGUUCGGGUAUCCUCAUUCAUCCUUGGCAUGCACGCACGUUUUUUCGAUCCGGCUCCUUUCUUUCCC ..(((((((((((((((-----((..(((((.((((.....))))..(((.(((((..(((.............)))..)))))))))))))....)))))...))))...)))))))). ( -42.22) >consensus GCGGAGAGAAGGUCUCG_____AGUGUGUGCGGUGGAACAUCCAUAAGUGUCAUGUUCGGGUAUCCUCAUUCAUCCUUGGCAUGCACGCACGUUUUUUCGAUCCGGCUCCUUUCUUUCCC ..((((((((((..(((.((((((..(((((.((((.....))))..(((.(((((..(((.............)))..)))))))))))))....)))))).)))..)).)))))))). (-39.82 = -41.82 + 2.00)

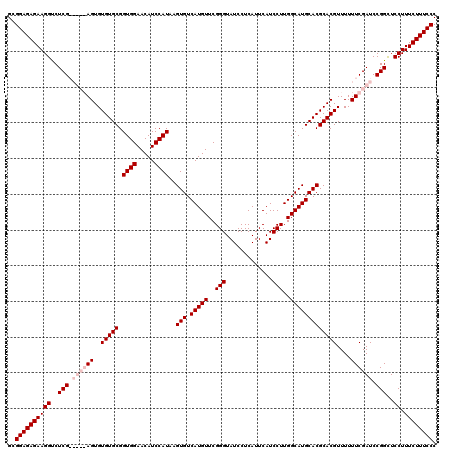
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:16 2006