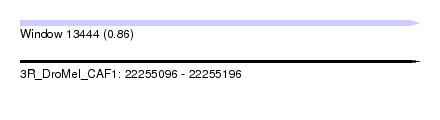
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,255,096 – 22,255,196 |
| Length | 100 |
| Max. P | 0.862594 |

| Location | 22,255,096 – 22,255,196 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.35 |
| Mean single sequence MFE | -30.15 |
| Consensus MFE | -16.15 |
| Energy contribution | -16.57 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
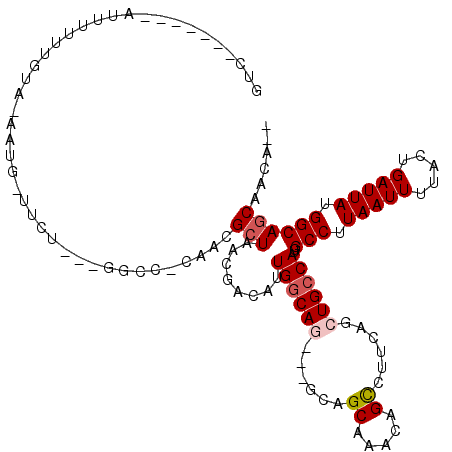
>3R_DroMel_CAF1 22255096 100 + 27905053 GUCUGCCAGGAUUUUUUGU--------------AGAC-CAACGCUAACGACAUUGGCAG---GCAGCAAACAGUUUUCAGCUGCCAUGGCCUUAAUUUAACUGAUUAUGGCAGCAACC-- (.((((((.((((..(((.--------------((.(-((..(((((.....))))).(---(((((............)))))).))).)).....)))..)))).)))))))....-- ( -30.60) >DroVir_CAF1 37246 104 + 1 GUC-------AAUUUUUGUAUAAUG-UUCU---GGCCCCAAUGGUAACGACAUUGGCAACGAACAGCUUUCUGCCUUCAGCUGCCAAGGCCUUAAUUUUACUGAUUAUGGCAGCA----- ...-------.............((-(((.---(...((((((.......))))))...))))))((.....)).....(((((((......(((((.....)))))))))))).----- ( -25.40) >DroGri_CAF1 56956 106 + 1 GGC---------UUUGUGUAUAAUG-UUCU---AGCC-GAAUGCUAACGAGAUUGGCAGCGAACAGCUUUCAGCCUUCAGCUGGCACGGCCUUAAUUUUACUGAUUAUGGCAGCAGCAGC .((---------(.(((((....((-(((.---.(((-((...(....)...)))))...))))).....((((.....)))))))))(((.(((((.....))))).))))))...... ( -30.20) >DroSec_CAF1 27742 100 + 1 GUCUGCCAGGAUUUUUUGU--------------GGAC-CAACGCUAACGACAUUGGCAG---GCAGCAAACAGUUUUCAGCUGCCAUGGCCUUAAUUUAACUGAUUAUGGCAGCAACA-- (((((((((..((.((.((--------------(...-...))).)).))..)))))))---))...............(((((((((..(...........)..)))))))))....-- ( -33.70) >DroMoj_CAF1 33650 106 + 1 GGC---------UUUUUGUAAAAUG-UUCU---GGCC-CAAUGCUAACGACAUUGGCAACGAGCAGCUUUCAGCCUUCAACUGCCACGGCCUUAAUUUUACUGAUUAUGGCAGCAACAGC (((---------(....((....((-(((.---.(((-.(((((....).)))))))...)))))))....))))......(((....(((.(((((.....))))).))).)))..... ( -27.60) >DroAna_CAF1 29840 110 + 1 GCCU----GAAUUUUUUGUUCUCAGUUUUUGGCGCUC-CAACGCUAACGACAUUGGCAG---GCAGCAAACAGUUUUCAGCUGCCAUGGCCUUAAUUUUACUGAUUAUGGCAGCAACC-- (((.----((((.....))))...(((.((((((...-...)))))).)))...))).(---(((((............))))))...(((.(((((.....))))).))).......-- ( -33.40) >consensus GUC_______AUUUUUUGUA_AAUG_UUCU___GGCC_CAACGCUAACGACAUUGGCAG___GCAGCAAACAGCCUUCAGCUGCCAUGGCCUUAAUUUUACUGAUUAUGGCAGCAACA__ ..........................................(((........((((((......((.....))......))))))..(((.(((((.....))))).))))))...... (-16.15 = -16.57 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:13 2006