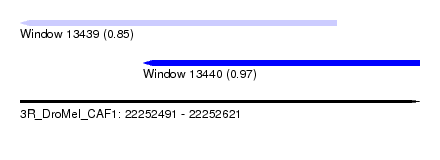
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,252,491 – 22,252,621 |
| Length | 130 |
| Max. P | 0.969222 |

| Location | 22,252,491 – 22,252,594 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.58 |
| Mean single sequence MFE | -22.74 |
| Consensus MFE | -8.79 |
| Energy contribution | -11.28 |
| Covariance contribution | 2.49 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22252491 103 - 27905053 UGGAAUCGGGGAAAACCCAGUUUCUGUACAUAAUUGUUUCAAGUGCUC---AUAAAGUGCUGAUAAAGGUUGGAAGAAACAGAUGCCAAAAUACUAAUUAAUUAUU (((.((((((.....))).((((((...((...((...(((.(..((.---....))..))))..))...))..)))))).))).))).................. ( -24.30) >DroSec_CAF1 25181 102 - 1 CGGAAUCGGGGAAAACCCAGAUUCUGUACAUAAUUGUUUCAAAUGCUC---AUA-AGUGCUGAUAAAGGUUGGAACACACAAAUGCCAAAAUACUGAUUAAUUAUU ((((((((((.....))).)))))))...(((((((..(((...((..---...-...)).......(((((.......))...))).......))).))))))). ( -19.80) >DroSim_CAF1 25305 102 - 1 CGGAAUCGGGGAAAACCCAGAUUCUGUACAUAAUUGUUUCAAGUGCUC---AUA-AGUGCUGAUAAAGGUUGGAACACACAAAUGCCAAAAUACUGAUUAAUUAUU ((((((((((.....))).)))))))...(((((((..(((((..((.---...-))..))......(((((.......))...))).......))).))))))). ( -23.00) >DroEre_CAF1 34286 79 - 1 ----AUCGGGGAAAUACCAGAUUCUGUACAUAAUUGUUUCCA-------------------GAUAAGUGCUGGAACCAAAAAAUUCUAAAAUACUGUAUA-GU--- ----(((.((......)).))).(((((((...(((.(((((-------------------(.......)))))).)))...(((....)))..))))))-).--- ( -15.20) >DroYak_CAF1 30188 104 - 1 UGGAAUCGGGGUAAGCCCAGAUUCUGUUCCUAAUUGUUUCAAGUGCUAAUCACA-AUUGCUGAUUAGUGCUGGAACAAACAAACACCAAAAUACUGAUUA-UUAUU .((((((.(((....))).))))))........((((((((.(..((((((((.-...).)))))))..)))))))))......................-..... ( -31.40) >consensus CGGAAUCGGGGAAAACCCAGAUUCUGUACAUAAUUGUUUCAAGUGCUC___AUA_AGUGCUGAUAAAGGUUGGAACAAACAAAUGCCAAAAUACUGAUUAAUUAUU (((((((.(((....))).)))))))........(((((((((((((........)))))).........)))))))............................. ( -8.79 = -11.28 + 2.49)



| Location | 22,252,531 – 22,252,621 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 88.52 |
| Mean single sequence MFE | -23.82 |
| Consensus MFE | -20.01 |
| Energy contribution | -20.82 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22252531 90 - 27905053 GCAUUUGAUUUACGUCACAAA-ACUGUUUGGAAUCGGGGAAAACCCAGUUUCUGUACAUAAUUGUUUCAAGUGCUC---AUAAAGUGCUGAUAA ((((((((........((((.-..(((..(((((.(((.....))).)))))...)))...)))).))))))))..---............... ( -19.90) >DroSec_CAF1 25221 89 - 1 GCAUUUGAUUUACGCCACAAA-ACUGUUCGGAAUCGGGGAAAACCCAGAUUCUGUACAUAAUUGUUUCAAAUGCUC---AUA-AGUGCUGAUAA ((((((((........((((.-..(((.((((((((((.....))).))))))).)))...)))).))))))))..---...-........... ( -25.30) >DroSim_CAF1 25345 89 - 1 GCAUUUGAUUUACGCCACAAA-UCUGUUCGGAAUCGGGGAAAACCCAGAUUCUGUACAUAAUUGUUUCAAGUGCUC---AUA-AGUGCUGAUAA ((((((((........((((.-(.(((.((((((((((.....))).))))))).))).).)))).))))))))..---...-........... ( -25.30) >DroYak_CAF1 30227 93 - 1 GCAUUUGAUUUACGCCCCAAAAGCUCUUUGGAAUCGGGGUAAGCCCAGAUUCUGUUCCUAAUUGUUUCAAGUGCUAAUCACA-AUUGCUGAUUA ((((((((...((((.......)).....((((((.(((....))).))))))..........)).))))))))(((((((.-...).)))))) ( -24.80) >consensus GCAUUUGAUUUACGCCACAAA_ACUGUUCGGAAUCGGGGAAAACCCAGAUUCUGUACAUAAUUGUUUCAAGUGCUC___AUA_AGUGCUGAUAA ((((((((........((((....(((.(((((((.(((....))).))))))).)))...)))).)))))))).................... (-20.01 = -20.82 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:09 2006