
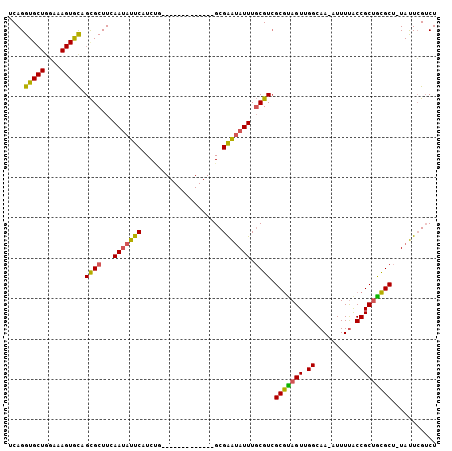
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,247,436 – 22,247,597 |
| Length | 161 |
| Max. P | 0.744644 |

| Location | 22,247,436 – 22,247,527 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -28.31 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22247436 91 + 27905053 UCAGGUGCUGGAAAGUGCAGCGCUUCAAUAUUCAUCUG-------------GCGAAUAUUUGCGUCGCGUAGUUGGCAA-AUUUUACCGCUGCGCU-UAUUCGUCU ..((((((((.......))))))))............(-------------(((((((...((((.(((.((((....)-)))....))).)))).-)))))))). ( -29.30) >DroSec_CAF1 19747 91 + 1 UCAGGUGCUGGAAAGUGCUGCGCUUCAAUAUUCAUCCG-------------GCGAAUAUUUGCGUCGCGUAGUUGGCAA-AUUUUACCGCUGCGCU-UAUUCGUCU ...((..((....))..))..................(-------------(((((((...((((.(((.((((....)-)))....))).)))).-)))))))). ( -26.90) >DroSim_CAF1 19827 91 + 1 UCAGGUGCUGGAAAGUGCUGCGCUUCAAUAUUCAUCCG-------------GCGAAUAUUUGCGUCGCGCAGUUGGCAA-AUUUUACCGCUGCGCU-UAUUCGUCU ...((..((....))..))((((...(((((((.....-------------..))))))).)))).(((((((.((...-......))))))))).-......... ( -28.60) >DroEre_CAF1 26833 91 + 1 UCAGGUGCUGGGAAGUGCAGCGCUUCAAAAUUCAUCUG-------------GCGAAUAUUUGCGUCGCGUAGUUGGCAA-AUUUUACCGCUGCGCU-UAUUCGGCU ......((((((((((((((((....((((((((.(((-------------((((.........)))).))).)))...-)))))..)))))))))-).)))))). ( -29.90) >DroYak_CAF1 24711 82 + 1 UCAGGU----------GCAGCGCUUCAAUAUUCAUCUG-------------GCGAGUAUUUGCGUCGCGUAGUUGGCAA-AUUUCACCGCUGCGCUUUACUCGUCU .(((((----------(...............))))))-------------(((((((...((((.(((.((((....)-)))....))).))))..))))))).. ( -23.86) >DroAna_CAF1 20987 105 + 1 UCAGGUGCUGCAAAGUAUAGUGCUUCAAUAUUCAUCGGAUGCUGGAAGCAAGCGGACAUUUUCGUGGCAUAGUUGGCAAAAUUUUGCCGCGAUGCU-UAUUGGUCU ..(((..(((.......)))..)))(((((..(((((..(((((....).))))..)......((((((.((((.....)))).))))))))))..-))))).... ( -31.30) >consensus UCAGGUGCUGGAAAGUGCAGCGCUUCAAUAUUCAUCUG_____________GCGAAUAUUUGCGUCGCGUAGUUGGCAA_AUUUUACCGCUGCGCU_UAUUCGUCU ....(((((....))))).((((...(((((((....................))))))).)))).(((((((.((..........)))))))))........... (-19.20 = -19.50 + 0.30)


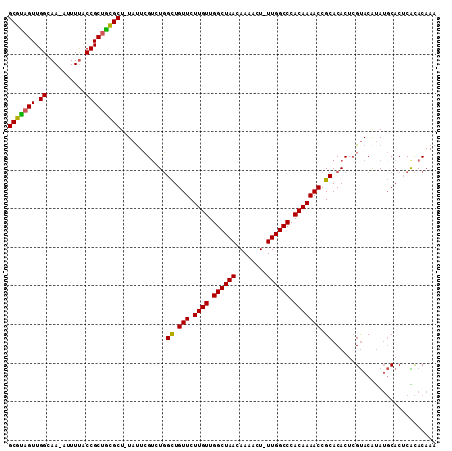
| Location | 22,247,489 – 22,247,597 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 91.74 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -24.74 |
| Energy contribution | -24.22 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22247489 108 + 27905053 GCGUAGUUGGCAA-AUUUUACCGCUGCGCU-UAUUCGUCUGGCUGUUCUUGUUGGCUAACAAAACU-UUGGCCCACAAAACCGCACACUCGUACGUAUGCACUCACACAAA (((((((.((...-......))))))))).-....((..((((.(((.((((.((((((.......-)))))).))))))).)).))..)).................... ( -26.90) >DroSec_CAF1 19800 108 + 1 GCGUAGUUGGCAA-AUUUUACCGCUGCGCU-UAUUCGUCUGGCUGUUCUUGUUGGCUAACAAAACU-UUGGCCCACAAAACCGCACACUCGUACGUAUGCACUCACACAAA (((((((.((...-......))))))))).-....((..((((.(((.((((.((((((.......-)))))).))))))).)).))..)).................... ( -26.90) >DroSim_CAF1 19880 108 + 1 GCGCAGUUGGCAA-AUUUUACCGCUGCGCU-UAUUCGUCUGGCUGUUCUUGUUGGCUAACAAAACU-UUGGCCCACAAAACCGCACACUCGUACGUAUGCACUCACACAAA (((((((.((...-......))))))))).-....((..((((.(((.((((.((((((.......-)))))).))))))).)).))..)).................... ( -28.90) >DroEre_CAF1 26886 108 + 1 GCGUAGUUGGCAA-AUUUUACCGCUGCGCU-UAUUCGGCUGGCUGUUCUUGUUGGCUAACAAAACU-UUGGCCCACAAAACCGCACACUAGUACAUAUGCACUCGCACAAG (((((((.((...-......))))))))).-((((.((.((((.(((.((((.((((((.......-)))))).))))))).)).))))))))....(((....))).... ( -27.50) >DroYak_CAF1 24754 109 + 1 GCGUAGUUGGCAA-AUUUCACCGCUGCGCUUUACUCGUCUGGCUGUUCUUGUUGGCUAACAAAACU-UUGGCCCACAAAACCGCACACUCGUACAUAUGCACUCACACAAG (((((((.((...-......)))))))))......((..((((.(((.((((.((((((.......-)))))).))))))).)).))..)).................... ( -26.90) >DroAna_CAF1 21053 108 + 1 GCAUAGUUGGCAAAAUUUUGCCGCGAUGCU-UAUUGGUCUUGUUGUUCUUGUUGGCUAACAAAACUUUUGGCCCACAAAACCGCACACUCGCACACA--CACGCGUACAAA ((((.((.(((((....))))))).)))).-..(((....(((.(((.((((.((((((........)))))).))))))).)))....(((.....--...)))..))). ( -27.70) >consensus GCGUAGUUGGCAA_AUUUUACCGCUGCGCU_UAUUCGUCUGGCUGUUCUUGUUGGCUAACAAAACU_UUGGCCCACAAAACCGCACACUCGUACAUAUGCACUCACACAAA (((((((.((..........)))))))))............((.(((.((((.((((((........)))))).))))))).))........................... (-24.74 = -24.22 + -0.53)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:08 2006