
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,245,506 – 22,245,666 |
| Length | 160 |
| Max. P | 0.771733 |

| Location | 22,245,506 – 22,245,616 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.75 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -20.30 |
| Energy contribution | -19.72 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22245506 110 + 27905053 AAACUUUAUUAACAUUUUUCGCAUGCUGCACGUGCAG----CGUUUGUGGCAGGAACGUCGC-GAC-GCGUCGCGUAUUUCGCGCUCUUUAAUGAAGUUUCAUAAAUUGUGAAAAU .(((((((((((.......((((((((((....))))----)))(((((((......)))))-)).-))).((((.....))))....)))))))))))((((.....)))).... ( -34.70) >DroVir_CAF1 21275 97 + 1 AAACUUUAUUAACAUUUUUCGCAUGCUGCACGUGCA-----CAUU---------GACAACGU-----GGGCCGCGUAUUUUGCGCUUUUUAAUGAAGUUUCAUAAAUUGUGAGAAU .(((((((((((............(((.(((((.(.-----....---------)...))))-----)))).((((.....))))...)))))))))))((((.....)))).... ( -22.00) >DroPse_CAF1 25393 116 + 1 AAACUUUAUUAACAUUUUUCGCAUGCUGCACGUGCAGCCCGCAUUUGUGGCAGGGACGACGGGGACGGCGGCGCGCGUUUCGCGCUCUUUAAUGAAGUUUCAUAAAUUGUGAAAAU .(((((((((((............(((((.(((.(...((((....))))...).))).((....)))))))(((((...)))))...)))))))))))((((.....)))).... ( -39.60) >DroGri_CAF1 46008 98 + 1 AAACUUUAUUAACAUUUUUCGCAUGCUGCACGUGCAG----CACU---------GACAACGU-----GGGCCGCGUAUUUUGUGCUUUUUAAUGAAGUUUCAUAAAUUGUGAGAAU .(((((((((((........(((((.....)))))((----(((.---------....((((-----(...))))).....)))))..)))))))))))((((.....)))).... ( -25.20) >DroAna_CAF1 18796 100 + 1 AAACUUUAUUAACAUUUUUCGCAUGCUGCACGUGCAG----CAUUUCUGCCAG-----------CC-GCCUUGCGUAUUUCGCGCUCUUUAAUGAAGUUUCAUAAAUUGUGAAAAU .(((((((((((........(((((((((....))))----))....)))...-----------..-.....((((.....))))...)))))))))))((((.....)))).... ( -24.90) >DroPer_CAF1 33378 116 + 1 AAACUUUAUUAACAUUUUUCGCAUGCUGCACGUGCAGCCCGCAUUUGUGGCAGGGACGACGGUGACGGCGGCGCGCGUUUCGCGCUCUUUAAUGAAGUUUCAUAAAUUGUGAAAAU .(((((((((((............(((((.(((.(...((((....))))...).))).((....)))))))(((((...)))))...)))))))))))((((.....)))).... ( -39.60) >consensus AAACUUUAUUAACAUUUUUCGCAUGCUGCACGUGCAG____CAUUUGUGGCAG_GACGACGG__AC_GCGCCGCGUAUUUCGCGCUCUUUAAUGAAGUUUCAUAAAUUGUGAAAAU ...(((((((((........(((((.....))))).....................................((((.....))))...)))))))))((((((.....)))))).. (-20.30 = -19.72 + -0.58)



| Location | 22,245,576 – 22,245,666 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.30 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -14.68 |
| Energy contribution | -15.02 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22245576 90 + 27905053 AUUUCGCGCUCUUUAAUGAAGUUUCAUAAAUUGUGAAAAUCGAACGUUAAUCAAAUAAGGAGCGG---------AGAGCCAUGUGCAAGAGGUGGCGCC .((((.(((((((((..((..((((((.....)))))).))((.......))...))))))))))---------)))(((((.(.....).)))))... ( -25.60) >DroVir_CAF1 21332 94 + 1 AUUUUGCGCUUUUUAAUGAAGUUUCAUAAAUUGUGAGAAUCGAACGUUAAUCAAAUAAGGUGCGAGAGAAAUCAAAAGCCUUGUCCG-----AGACGCC .....((((((.((((((...((((((.....))))))......))))))....(((((((....((....))....)))))))..)-----)).))). ( -19.40) >DroGri_CAF1 46066 94 + 1 AUUUUGUGCUUUUUAAUGAAGUUUCAUAAAUUGUGAGAAUCGAACGUUAAUCAAAUAAGGUGCGAGAGAAAUCAAAAGCCUUGUCCC-----AGGCGCC .....((((((.((((((...((((((.....))))))......))))))....(((((((....((....))....)))))))...-----)))))). ( -20.60) >DroEre_CAF1 25005 90 + 1 AUUUUGCGCUCUUUAAUGAAGUUUCAUAAAUUGUGAAAAUCGAACGUUAAUCAAAUAAGGAGCGG---------AAAGCCAUGUACAAAAGGUGGCCCC ..(((.(((((((((..((..((((((.....)))))).))((.......))...))))))))).---------)))(((((.(.....).)))))... ( -23.40) >DroYak_CAF1 22884 90 + 1 AUUUCGCGCUCUUUAAUGAAGUUUCAUAAAUUGUGAAAAUCGAACGUUAAUCAAAUAAGGAGCGG---------AGAGCCAUGUACACAAGGUGGCGCC .((((.(((((((((..((..((((((.....)))))).))((.......))...))))))))))---------)))(((((.(.....).)))))... ( -25.40) >DroAna_CAF1 18856 86 + 1 AUUUCGCGCUCUUUAAUGAAGUUUCAUAAAUUGUGAAAAUCGAACGUUAAUCAAAUAAGGAGCGG---------GG---CCCGCACA-AAGGUGGCGCC ......(((((((((..((..((((((.....)))))).))((.......))...))))))))).---------((---(((((...-...)))).))) ( -24.50) >consensus AUUUCGCGCUCUUUAAUGAAGUUUCAUAAAUUGUGAAAAUCGAACGUUAAUCAAAUAAGGAGCGG_________AAAGCCAUGUACA_AAGGUGGCGCC ......(((((((((..((..((((((.....)))))).))((.......))...))))))))).............(((((.........)))))... (-14.68 = -15.02 + 0.34)

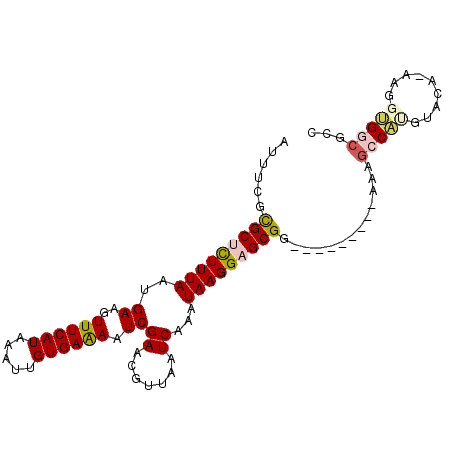
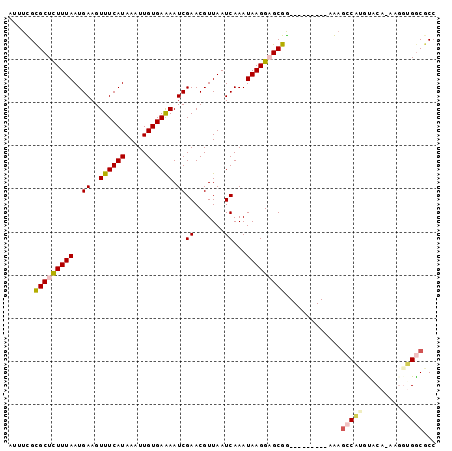
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:09:04 2006