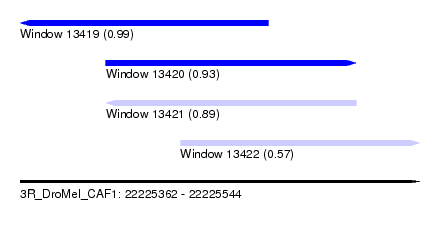
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,225,362 – 22,225,544 |
| Length | 182 |
| Max. P | 0.993676 |

| Location | 22,225,362 – 22,225,475 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 88.66 |
| Mean single sequence MFE | -35.55 |
| Consensus MFE | -31.92 |
| Energy contribution | -32.93 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22225362 113 - 27905053 GCUGCCGGCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUUCGCCGGCUGACAGCCACACUCA-CACUAUGACGCACACACACACACAC ((.((((((..(((((.(((((.(((((((((..........))).)))))))))))...)))))..))))))............(((-.....))).)).............. ( -37.80) >DroSec_CAF1 68175 111 - 1 GCUGCCGGCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUUCGCCGGCUGACAGCCACACUCA-CACUAUGACACACAC--ACACACAC (((((((((..(((((.(((((.(((((((((..........))).)))))))))))...)))))..))))))....)))........-...............--........ ( -37.70) >DroSim_CAF1 59699 113 - 1 GCUGCCGGCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUUCGCCGGCUGACAGCCACACUCA-CACUAUGACACACACACACACACAC (((((((((..(((((.(((((.(((((((((..........))).)))))))))))...)))))..))))))....)))........-......................... ( -37.70) >DroAna_CAF1 93688 95 - 1 GCUGCCGGCAUGUGGUCGGUUGACCGGGCCAA--AACCAGAAUUGC---CGGGAACUUAAAUCAUUCGCCGGCUGACAGCCACACACAGCACU--GAUAUAC------------ ((((((((((...((((....))))((.....--..)).....)))---)))..................(((.....))).....))))...--.......------------ ( -29.00) >consensus GCUGCCGGCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUUCGCCGGCUGACAGCCACACUCA_CACUAUGACACACAC__ACACACAC (((((((((..(((((.(((((.(((((((((..........))).)))))))))))...)))))..))))))....))).................................. (-31.92 = -32.93 + 1.00)

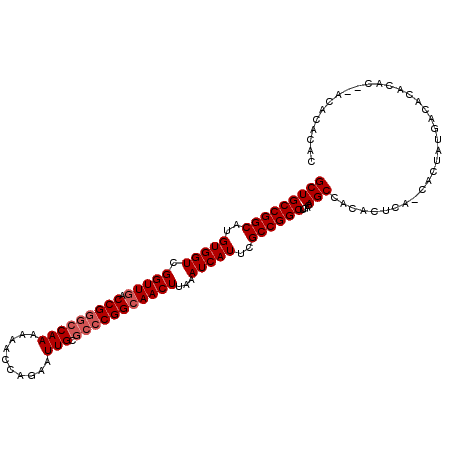
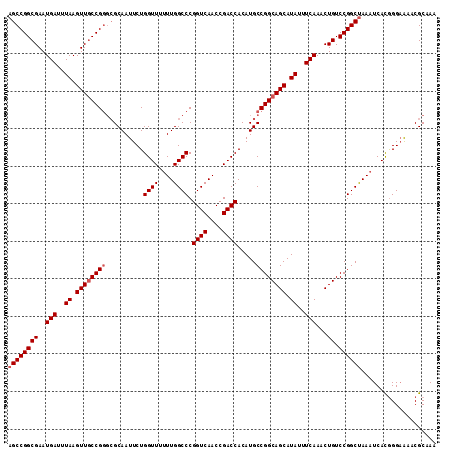
| Location | 22,225,401 – 22,225,515 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 95.18 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -34.35 |
| Energy contribution | -35.10 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22225401 114 + 27905053 AGCCGGCGAAUGAUUUAAGUUGCCGGGCGCAAUUCUGGUUUUUUGGCCCGGUCAACCGACCACAUGCCGGCAGCAUAUUUCAAACUGUCCGGCUAAAUCACGGGAAAACGCAAA ((((((((..(((..((.((((((((((.(((..........)))))))((((....)))).......)))))).))..)))...)).))))))......((......)).... ( -40.60) >DroSec_CAF1 68212 114 + 1 AGCCGGCGAAUGAUUUAAGUUGCCGGGCGCAAUUCUGGUUUUUUGGCCCGGUCAACCGACCACAUGCCGGCAGCAUAUUUCAAACUGUCCGGCUAAAUCAUGGGAAAACGCAAA ((((((((..(((..((.((((((((((.(((..........)))))))((((....)))).......)))))).))..)))...)).)))))).................... ( -40.30) >DroSim_CAF1 59738 114 + 1 AGCCGGCGAAUGAUUUAAGUUGCCGGGCGCAAUUCUGGUUUUUUGGCCCGGUCAACCGACCACAUGCCGGCAGCAUAUUUCAAACUGUCCGGCUAAAUCACGGGAAAACGCAAA ((((((((..(((..((.((((((((((.(((..........)))))))((((....)))).......)))))).))..)))...)).))))))......((......)).... ( -40.60) >DroAna_CAF1 93714 109 + 1 AGCCGGCGAAUGAUUUAAGUUCCCG---GCAAUUCUGGUU--UUGGCCCGGUCAACCGACCACAUGCCGGCAGCAUAUUUCAAACUGUCCGGCCAAAUCACAGGCGAACGCAAA .(((((.((((.......)))))))---))....(((..(--((((((.((((....)))).......(((((...........))))).)))))))...)))((....))... ( -39.30) >consensus AGCCGGCGAAUGAUUUAAGUUGCCGGGCGCAAUUCUGGUUUUUUGGCCCGGUCAACCGACCACAUGCCGGCAGCAUAUUUCAAACUGUCCGGCUAAAUCACGGGAAAACGCAAA ((((((((..(((..((.((((((((..........((((....)))).((((....)))).....)))))))).))..)))...)).)))))).................... (-34.35 = -35.10 + 0.75)

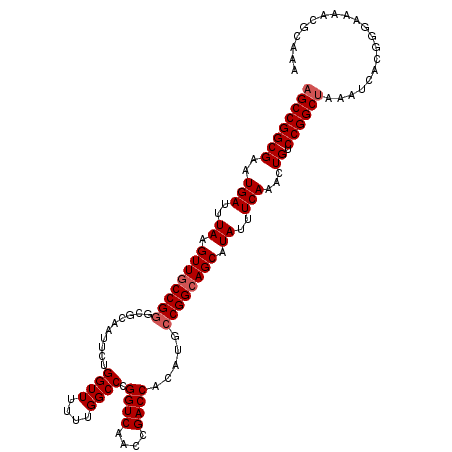
| Location | 22,225,401 – 22,225,515 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 95.18 |
| Mean single sequence MFE | -41.00 |
| Consensus MFE | -35.67 |
| Energy contribution | -36.30 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
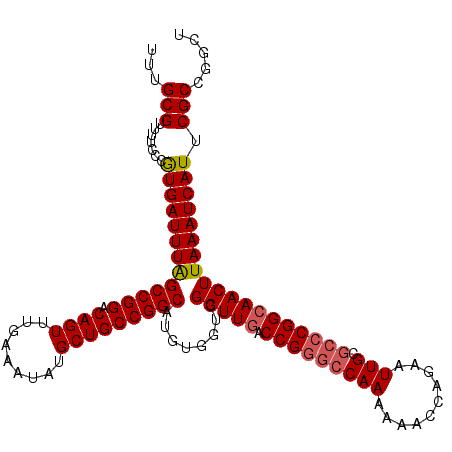
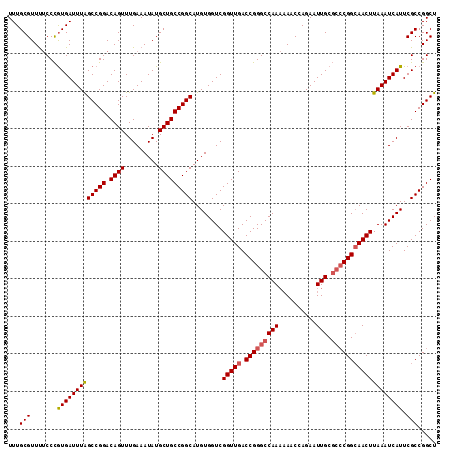
>3R_DroMel_CAF1 22225401 114 - 27905053 UUUGCGUUUUCCCGUGAUUUAGCCGGACAGUUUGAAAUAUGCUGCCGGCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUUCGCCGGCU .(..((......))..)...((((((.((((.........))))))(((..(((((.(((((.(((((((((..........))).)))))))))))...)))))..))))))) ( -42.50) >DroSec_CAF1 68212 114 - 1 UUUGCGUUUUCCCAUGAUUUAGCCGGACAGUUUGAAAUAUGCUGCCGGCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUUCGCCGGCU ...(((.......(((((((((((((.((((.........)))))))))........(((((.(((((((((..........))).))))))))))))))))))).)))..... ( -40.80) >DroSim_CAF1 59738 114 - 1 UUUGCGUUUUCCCGUGAUUUAGCCGGACAGUUUGAAAUAUGCUGCCGGCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUUCGCCGGCU .(..((......))..)...((((((.((((.........))))))(((..(((((.(((((.(((((((((..........))).)))))))))))...)))))..))))))) ( -42.50) >DroAna_CAF1 93714 109 - 1 UUUGCGUUCGCCUGUGAUUUGGCCGGACAGUUUGAAAUAUGCUGCCGGCAUGUGGUCGGUUGACCGGGCCAA--AACCAGAAUUGC---CGGGAACUUAAAUCAUUCGCCGGCU ((((.(((.((((........(((((.((((.........)))))))))....((((....))))))))...--)))))))...((---((((((.........))).))))). ( -38.20) >consensus UUUGCGUUUUCCCGUGAUUUAGCCGGACAGUUUGAAAUAUGCUGCCGGCAUGUGGUCGGUUGACCGGGCCAAAAAACCAGAAUUGCGCCCGGCAACUUAAAUCAUUCGCCGGCU ...(((.......(((((((((((((.((((.........)))))))))........(((((.(((((((((..........))).))))))))))))))))))).)))..... (-35.67 = -36.30 + 0.63)

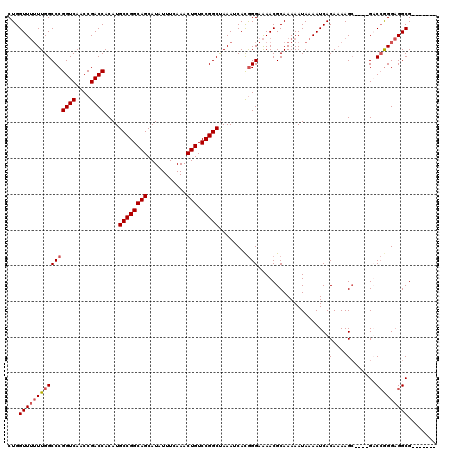
| Location | 22,225,435 – 22,225,544 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.37 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -24.01 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22225435 109 + 27905053 CUGGUUUUUUGGCCCGGUCAACCGACCACAUGCCGGCAGCAUAUUUCAAACUGUCCGGCUAAAUCACGGGAAAACGCAAAAAUAAAAUCACAAAAGC----GUCCGGGAGGCG------- ...((((((((((((((((....))))....((((((((...........))).)))))........)))...((((..................))----))))))))))).------- ( -33.47) >DroSec_CAF1 68246 109 + 1 CUGGUUUUUUGGCCCGGUCAACCGACCACAUGCCGGCAGCAUAUUUCAAACUGUCCGGCUAAAUCAUGGGAAAACGCAAAAAUAAAAUCACAAAAGC----GACCGGGAGGCG------- ...((((((((((((((((....))))....((((((((...........))).)))))........)))....(((..................))----).))))))))).------- ( -33.07) >DroSim_CAF1 59772 109 + 1 CUGGUUUUUUGGCCCGGUCAACCGACCACAUGCCGGCAGCAUAUUUCAAACUGUCCGGCUAAAUCACGGGAAAACGCAAAAAUAAAAUCACAAAAGC----GACCGGGAGGCG------- ...((((((((((((((((....))))....((((((((...........))).)))))........)))....(((..................))----).))))))))).------- ( -33.37) >DroAna_CAF1 93745 118 + 1 CUGGUU--UUGGCCCGGUCAACCGACCACAUGCCGGCAGCAUAUUUCAAACUGUCCGGCCAAAUCACAGGCGAACGCAAAAAUAAAAUCACAUAAGAAGGAGUCGAGGUGGCGGGGUGGC ......--...((((.((((.(((((..(..((((((((...........))).)))))..........((....)).....................)..)))).).)))).))))... ( -33.60) >consensus CUGGUUUUUUGGCCCGGUCAACCGACCACAUGCCGGCAGCAUAUUUCAAACUGUCCGGCUAAAUCACGGGAAAACGCAAAAAUAAAAUCACAAAAGC____GACCGGGAGGCG_______ ...((((((((((((((((....))))....((((((((...........))).)))))........)))....................(....).......)))))))))........ (-24.01 = -24.82 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:53 2006