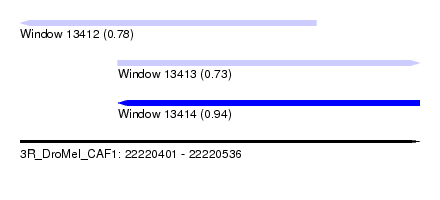
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,220,401 – 22,220,536 |
| Length | 135 |
| Max. P | 0.940599 |

| Location | 22,220,401 – 22,220,501 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.06 |
| Mean single sequence MFE | -35.37 |
| Consensus MFE | -20.26 |
| Energy contribution | -22.35 |
| Covariance contribution | 2.09 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780436 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22220401 100 - 27905053 GUGCCGCUGUCAAUAAGGAUUGGCAGUCAGCAUGCGGGUUGGUUCAGACUUGG--GGGCCCG------CCUUUCUCCAUUCGCUGCCAAUCCCUGGCCAUAUUUUCUU-- (.((((((((((((....)))))))))........((((((((...((..(((--(((....------....)))))).))...))))))))..))))..........-- ( -34.60) >DroSec_CAF1 55798 100 - 1 GUGCCGCUGUCAAUAAGGAUUGGCAGUCAGCAUGCGGGUUGGUUCAGACUUGG--GGGCACG------CCUUUCCCCUUUCACUGCCAAUCCCCGGCCAUAUUUUCUU-- (.((((((((((((....)))))))))......(.((((((((...((...((--(((....------....)))))..))...)))))))).)))))..........-- ( -35.30) >DroSim_CAF1 54733 100 - 1 GUGCCGCUGUCAAUAAGGAUUGGCAGUCAGCAUGCGGGUUGGUUCAGACUUGG--GGGCACG------CCUUUCCCCUUUCGCUGCCAAUCCCCGGCCAUAUUUUCUU-- (.((((((((((((....)))))))))......(.((((((((...((...((--(((....------....)))))..))...)))))))).)))))..........-- ( -36.40) >DroEre_CAF1 69969 101 - 1 GUGCCGCUGUCAAUAAGGAUUGGCAGUCAGCAUGCGGGUUGGCUCAGACUUGU--GGGCCCG------CCUUUACCCUCUCGCUGCCAAUCCUGG-CCAUAUUUUUUUUU (.(((..........((((((((((((.((..((.((((.((((((......)--))))).)------))).))....)).))))))))))))))-))............ ( -37.80) >DroYak_CAF1 63020 101 - 1 GUGCCGCUGUCAAUAAGGAUUGGCAGUCAGCAUGCGGGUUGGUUCAGACUUGG--GGUCCCG------CCUUUCCCCUUUCGCUGCCAAUCCCUG-GUUCAAUUUUUUUA ((((.(((((((((....)))))))))..))))..((((((((...((...((--((.....------.....))))..))...))))))))...-.............. ( -34.10) >DroMoj_CAF1 80177 91 - 1 GUGCUGCUGUCAAUGAGGAUUGGCAGGCAGCAUGCGGGCUGAGCAGGACUGGGCU-----CGGAUCAGCUUUUGGCUUUUGG------------GGCUUUCAUUUCUU-- ((((((((((((((....))))))).))))))).((((((.((.....)).))))-----))((..((((((........))------------)))).)).......-- ( -34.00) >consensus GUGCCGCUGUCAAUAAGGAUUGGCAGUCAGCAUGCGGGUUGGUUCAGACUUGG__GGGCCCG______CCUUUCCCCUUUCGCUGCCAAUCCCCGGCCAUAUUUUCUU__ ((((.(((((((((....)))))))).).))))..((((((((...((...((..(((..........)))....))..))...)))))))).................. (-20.26 = -22.35 + 2.09)


| Location | 22,220,434 – 22,220,536 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.84 |
| Mean single sequence MFE | -30.89 |
| Consensus MFE | -20.77 |
| Energy contribution | -20.77 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22220434 102 + 27905053 AGAAAGG------CGGGCCC----CCAAGUCUGAACCAACCCGCAUGCUGACUGCCAAUCCUUAUUGACAGCGGCACUUCCGCUUUGGCAGCCAAAAUAGCUGCGAGCCCCA .(.((((------((((...----...............))))).(((((.(((.((((....)))).))))))))))).)((((..(((((.......))))))))).... ( -30.97) >DroVir_CAF1 89151 112 + 1 CCAAAAGCUGAGCCGUGAACUCAGCCCAGACCAGCUCAGCCCGCAUGCUGCCUGCCAAUCCUUAUUGACAGCAGCACUUCCGCUUUGGCAGCCAAAAUAGCUGCGAGCGCCA ......((((((.......))))))........(((((((.....((((((.((.((((....)))).)))))))).....)))...(((((.......))))))))).... ( -37.10) >DroEre_CAF1 70003 102 + 1 GUAAAGG------CGGGCCC----ACAAGUCUGAGCCAACCCGCAUGCUGACUGCCAAUCCUUAUUGACAGCGGCACUUCCGCCUUGGCAGCCAAAAUAGCUGCGAGCCCCA ...((((------(((((.(----(......)).)))........(((((.(((.((((....)))).))))))))....)))))).(((((.......)))))........ ( -33.60) >DroYak_CAF1 63054 102 + 1 GGAAAGG------CGGGACC----CCAAGUCUGAACCAACCCGCAUGCUGACUGCCAAUCCUUAUUGACAGCGGCACUUCCGCUUUGGCAGCCAAAAUAGCUGCGAGCCCCA ((((..(------((((...----...............))))).(((((.(((.((((....)))).)))))))).))))((((..(((((.......))))))))).... ( -34.07) >DroMoj_CAF1 80198 105 + 1 CCAAAAGCUGAUCCG-------AGCCCAGUCCUGCUCAGCCCGCAUGCUGCCUGCCAAUCCUCAUUGACAGCAGCACUUCCGCUUUGGCAGCCAAAAUAGCUGCGAGCGCCA ......(((((.(.(-------..(...)..).).)))))..((.((((((.((.((((....)))).)))))))).....((((..(((((.......))))))))))).. ( -31.30) >DroAna_CAF1 88923 85 + 1 ---------------------------AGCCUGAACUAACCCGCAUGCUGACUGCCAAUCCUUAUUGACAGCCGCACUUCCGCAUUGGCAGCCAAAAUAGCUGCGAGCCCCA ---------------------------..............((((.((((.((((((((.(.....)......((......))))))))))......))))))))....... ( -18.30) >consensus CCAAAGG______CGGG_CC____CCAAGUCUGAACCAACCCGCAUGCUGACUGCCAAUCCUUAUUGACAGCGGCACUUCCGCUUUGGCAGCCAAAAUAGCUGCGAGCCCCA ..........................................((.(((((.(((.((((....)))).))))))))...........(((((.......)))))..)).... (-20.77 = -20.77 + 0.00)



| Location | 22,220,434 – 22,220,536 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.84 |
| Mean single sequence MFE | -43.00 |
| Consensus MFE | -31.73 |
| Energy contribution | -30.98 |
| Covariance contribution | -0.74 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22220434 102 - 27905053 UGGGGCUCGCAGCUAUUUUGGCUGCCAAAGCGGAAGUGCCGCUGUCAAUAAGGAUUGGCAGUCAGCAUGCGGGUUGGUUCAGACUUGG----GGGCCCG------CCUUUCU ..((((((.(.((.((.((((((((((((((((.....))))).((......))))))))))))).))))(((((......)))))).----)))))).------....... ( -41.80) >DroVir_CAF1 89151 112 - 1 UGGCGCUCGCAGCUAUUUUGGCUGCCAAAGCGGAAGUGCUGCUGUCAAUAAGGAUUGGCAGGCAGCAUGCGGGCUGAGCUGGUCUGGGCUGAGUUCACGGCUCAGCUUUUGG ..(((((.((((((.....))))))...)))....((((((((((((((....))))))).)))))))))(((((((((((...(((((...)))))))))))))))).... ( -51.50) >DroEre_CAF1 70003 102 - 1 UGGGGCUCGCAGCUAUUUUGGCUGCCAAGGCGGAAGUGCCGCUGUCAAUAAGGAUUGGCAGUCAGCAUGCGGGUUGGCUCAGACUUGU----GGGCCCG------CCUUUAC ..(((((((((((.((.((((((((((((((((.....))))).((......))))))))))))).))))(((....))).....)))----)))))).------....... ( -45.10) >DroYak_CAF1 63054 102 - 1 UGGGGCUCGCAGCUAUUUUGGCUGCCAAAGCGGAAGUGCCGCUGUCAAUAAGGAUUGGCAGUCAGCAUGCGGGUUGGUUCAGACUUGG----GGUCCCG------CCUUUCC .(((((((((((((.....((((((((((((((.....))))).((......)))))))))))))).)))(((((......))))).)----)))))).------....... ( -41.80) >DroMoj_CAF1 80198 105 - 1 UGGCGCUCGCAGCUAUUUUGGCUGCCAAAGCGGAAGUGCUGCUGUCAAUGAGGAUUGGCAGGCAGCAUGCGGGCUGAGCAGGACUGGGCU-------CGGAUCAGCUUUUGG ...((((.((((((.....))))))...))))(((((((((((((((((....))))))).)))))((.((((((.((.....)).))))-------)).))..)))))... ( -44.50) >DroAna_CAF1 88923 85 - 1 UGGGGCUCGCAGCUAUUUUGGCUGCCAAUGCGGAAGUGCGGCUGUCAAUAAGGAUUGGCAGUCAGCAUGCGGGUUAGUUCAGGCU--------------------------- ..(((((.((((((.....)))))).(((.((...((((((((((((((....)))))))))).)))).)).)))))))).....--------------------------- ( -33.30) >consensus UGGGGCUCGCAGCUAUUUUGGCUGCCAAAGCGGAAGUGCCGCUGUCAAUAAGGAUUGGCAGUCAGCAUGCGGGUUGGUUCAGACUUGG____GG_CCCG______CCUUUCC ((((((((((((((.....((((((((((((((.....))))).((......)))))))))))))).)))))))...))))............................... (-31.73 = -30.98 + -0.74)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:46 2006