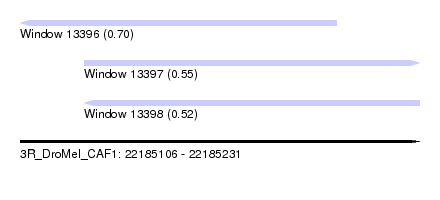
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,185,106 – 22,185,231 |
| Length | 125 |
| Max. P | 0.696592 |

| Location | 22,185,106 – 22,185,205 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.36 |
| Mean single sequence MFE | -31.46 |
| Consensus MFE | -18.82 |
| Energy contribution | -20.13 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22185106 99 - 27905053 -UAAUUGGCCUCAUUAGAUUUGAUGGCCCAGAAAGGGAUGUGUGCGUGAGUGCGGGUCUUGGCCAAACUAUGUGGCGCAGAGUGGAUUUG---------------AAUUCACCUG -.....(((((((.......))).))))......(((((.((..(....)..)).))))).((((.......)))).(((.((((((...---------------.))))))))) ( -33.70) >DroPse_CAF1 55943 104 - 1 UUAAUUGGCCUCAUUAGAUUUAAUGGC--UGAAUGGUAUGUGUGUUUGC--------UCGGAGGAAACGUUGUUUCGCCGA-UGGCCUUGUGAUAAAAGAAGGCGAGAACACCUU ....(..(((..............)))--..).........((((((..--------((((..(((((...))))).))))-..(((((.(......).)))))..))))))... ( -27.34) >DroSec_CAF1 29630 99 - 1 -UAAUUGGCCUCAUUAGAUUUGAUGGCCCAGAAAGAGAUGUGUGCGUGAGUGCGGGUUUUGGCCAAACUAUGUGGCGCAGAGUGGAUUUG---------------AAUUCACCUG -.....(((((((.......))).))))......(((((.((..(....)..)).))))).((((.......)))).(((.((((((...---------------.))))))))) ( -31.90) >DroSim_CAF1 28462 99 - 1 -UAAUUGGCCUCAUUAGAUUUGAUGGCCCAGAAAGGGAUGUGUGCGUGAGUGCGGGUUUUGGCCAAACUAUGUGGUGCAGAGUGGAUUUG---------------GAUUCACCUG -...((((((((((((....))))))(((.....)))((.((..(....)..)).))...))))))...........(((.((((((...---------------.))))))))) ( -29.60) >DroEre_CAF1 43817 99 - 1 -UAAUUGGCCUCAUUAGAUUUGAUGGCCCAGAAAAGGAUGUGCGCGUGAGUGUGGGUUCUGGCCAAACUAUGUUGCGUAGGGUGGAUGUG---------------AAUUCACCUG -...((((((((((((....))))))........(((((.(((((....))))).))))))))))).(((((...)))))(((((((...---------------.))))))).. ( -35.20) >DroYak_CAF1 35946 99 - 1 -UAAUUGGCCUCAUUAGAUUUGAUGGCCCAGAAAAGGAUGUGUGCGUGGGUGUGGAUUCUGGCCAAACUAUGUUGCGCAGGGUGGAUUUG---------------AAUUCACCUG -.....(((((((.......))).))))..........(((((((((((...(((.......)))..))))).))))))((((((((...---------------.)))))))). ( -31.00) >consensus _UAAUUGGCCUCAUUAGAUUUGAUGGCCCAGAAAGGGAUGUGUGCGUGAGUGCGGGUUCUGGCCAAACUAUGUGGCGCAGAGUGGAUUUG_______________AAUUCACCUG ....((((((((((((....))))))........(((((.(((((....))))).)))))))))))...........(((.(((((((.................)))))))))) (-18.82 = -20.13 + 1.31)

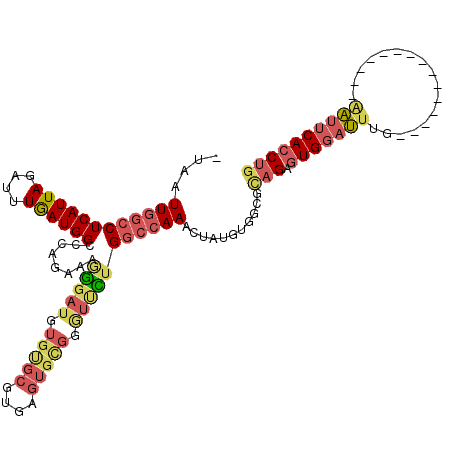

| Location | 22,185,126 – 22,185,231 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -20.20 |
| Consensus MFE | -14.41 |
| Energy contribution | -15.00 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22185126 105 + 27905053 CUGCGCCACAUAGUUUGGCCAAGACCCGCACUCACGCACACAUCCCUUUCUGGGCCAUCAAAUCUAAUGAGGCCAAUUA-AACCACACACUCUAGUGUUGGCCCCA ....((((......((((((.......((......))......(((.....)))................))))))...-......((((....)))))))).... ( -23.60) >DroPse_CAF1 55977 94 + 1 CGGCGAAACAACGUUUCCUCCGA--------GCAAACACACAUACCAUUCA--GCCAUUAAAUCUAAUGAGGCCAAUUAAAACCACAUACUCU--UACUCUACGCA (((.(((((...)))))..))).--------((..................--..(((((....))))).((..........)).........--........)). ( -9.50) >DroSec_CAF1 29650 105 + 1 CUGCGCCACAUAGUUUGGCCAAAACCCGCACUCACGCACACAUCUCUUUCUGGGCCAUCAAAUCUAAUGAGGCCAAUUA-AACCACACACUCCAGUGUUGGCCCCA .((.(((.(((((..(((((.......((......))...((........)))))))......)).))).)))))....-..(((.((((....)))))))..... ( -22.80) >DroSim_CAF1 28482 105 + 1 CUGCACCACAUAGUUUGGCCAAAACCCGCACUCACGCACACAUCCCUUUCUGGGCCAUCAAAUCUAAUGAGGCCAAUUA-AACCACACACUCCAGUGUUGGCCCCA ..............((((((.......((......))......(((.....)))................))))))...-..(((.((((....)))))))..... ( -21.60) >DroEre_CAF1 43837 105 + 1 CUACGCAACAUAGUUUGGCCAGAACCCACACUCACGCGCACAUCCUUUUCUGGGCCAUCAAAUCUAAUGAGGCCAAUUA-AACCACACACUCCAGUAUUGGCCCCA ..........(((..(((((((((.......................)))).)))))......))).((.(((((((..-................))))))).)) ( -18.37) >DroYak_CAF1 35966 105 + 1 CUGCGCAACAUAGUUUGGCCAGAAUCCACACCCACGCACACAUCCUUUUCUGGGCCAUCAAAUCUAAUGAGGCCAAUUA-AGCCACACACUCCAGUGUUGGCCCCA ..((.(((((((((.(((((((((.......................)))))((((.(((.......))))))).....-.))))...)))...)))))))).... ( -25.30) >consensus CUGCGCAACAUAGUUUGGCCAAAACCCGCACUCACGCACACAUCCCUUUCUGGGCCAUCAAAUCUAAUGAGGCCAAUUA_AACCACACACUCCAGUGUUGGCCCCA ................((((((..............................((((.(((.......))))))).............(((....)))))))))... (-14.41 = -15.00 + 0.59)


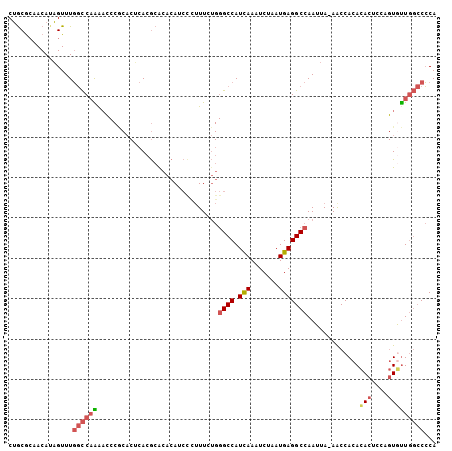
| Location | 22,185,126 – 22,185,231 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -33.51 |
| Consensus MFE | -23.81 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22185126 105 - 27905053 UGGGGCCAACACUAGAGUGUGUGGUU-UAAUUGGCCUCAUUAGAUUUGAUGGCCCAGAAAGGGAUGUGUGCGUGAGUGCGGGUCUUGGCCAAACUAUGUGGCGCAG ..(.((((((((....))))((((((-(....(((((((.......))).))))......(((((.((..(....)..)).)))))....))))))).)))).).. ( -38.00) >DroPse_CAF1 55977 94 - 1 UGCGUAGAGUA--AGAGUAUGUGGUUUUAAUUGGCCUCAUUAGAUUUAAUGGC--UGAAUGGUAUGUGUGUUUGC--------UCGGAGGAAACGUUGUUUCGCCG ......(((((--((..((((((.(.....(..(((..............)))--..)..).))))))..)))))--------))((..(((((...))))).)). ( -21.54) >DroSec_CAF1 29650 105 - 1 UGGGGCCAACACUGGAGUGUGUGGUU-UAAUUGGCCUCAUUAGAUUUGAUGGCCCAGAAAGAGAUGUGUGCGUGAGUGCGGGUUUUGGCCAAACUAUGUGGCGCAG ..(.((((((((....))))((((((-(....(((((((.......))).))))......(((((.((..(....)..)).)))))....))))))).)))).).. ( -36.20) >DroSim_CAF1 28482 105 - 1 UGGGGCCAACACUGGAGUGUGUGGUU-UAAUUGGCCUCAUUAGAUUUGAUGGCCCAGAAAGGGAUGUGUGCGUGAGUGCGGGUUUUGGCCAAACUAUGUGGUGCAG ..(.((((((((....))))((((((-....(((((((((((....))))))(((.....)))((.((..(....)..)).))...))))))))))).)))).).. ( -34.10) >DroEre_CAF1 43837 105 - 1 UGGGGCCAAUACUGGAGUGUGUGGUU-UAAUUGGCCUCAUUAGAUUUGAUGGCCCAGAAAAGGAUGUGCGCGUGAGUGUGGGUUCUGGCCAAACUAUGUUGCGUAG ((((((((((..((((.(....).))-))))))))))))..........(((((......(((((.(((((....))))).))))))))))..(((((...))))) ( -35.60) >DroYak_CAF1 35966 105 - 1 UGGGGCCAACACUGGAGUGUGUGGCU-UAAUUGGCCUCAUUAGAUUUGAUGGCCCAGAAAAGGAUGUGUGCGUGGGUGUGGAUUCUGGCCAAACUAUGUUGCGCAG ..((((((((((....))))(.((((-.....)))).)...........)))))).........(((((((((((...(((.......)))..))))).)))))). ( -35.60) >consensus UGGGGCCAACACUGGAGUGUGUGGUU_UAAUUGGCCUCAUUAGAUUUGAUGGCCCAGAAAGGGAUGUGUGCGUGAGUGCGGGUUCUGGCCAAACUAUGUGGCGCAG ..((((((.((.......(((.((((......)))).)))......)).)))))).....(((((.(((((....))))).))))).((((.......)))).... (-23.81 = -24.40 + 0.59)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:30 2006