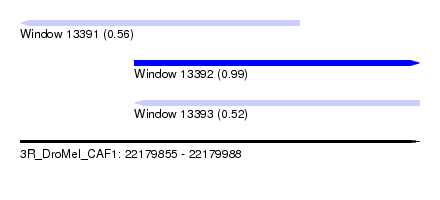
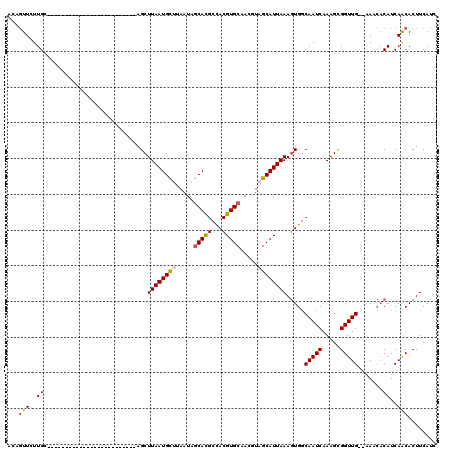
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,179,855 – 22,179,988 |
| Length | 133 |
| Max. P | 0.985251 |

| Location | 22,179,855 – 22,179,948 |
|---|---|
| Length | 93 |
| Sequences | 6 |
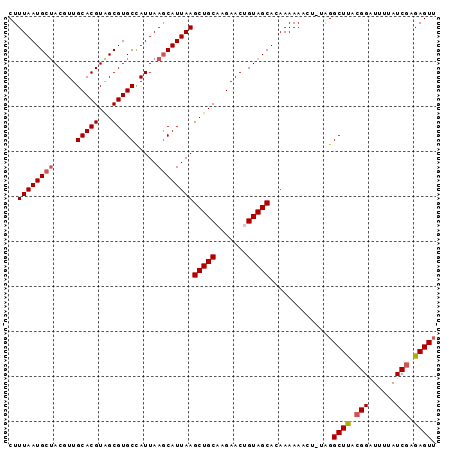
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.14 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -15.31 |
| Energy contribution | -15.57 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22179855 93 - 27905053 ACCGUUCUUGC-------------------------AGCUUAAUGUUUAAUGGCACGCUACGUGCAACGUAGCAUUAAAGUGGCAAUCAAAGCGGUUG--AAACACAUCAACACUUCAUC ..((((.((((-------------------------.((((((((((.....(((((...))))).....)))))).)))).))))....))))((((--(......)))))........ ( -25.10) >DroSec_CAF1 24369 93 - 1 ACAGUUCUUGC-------------------------AGCUUAAUGCUUAAUGGCACGCCACGUGCAACGUAGCAUUAAAGUGGCAAUCAAAGCGGUUG--AAACACAUCAACACUUCAUC ...(((.((((-------------------------.((((((((((.....(((((...))))).....)))))).)))).))))....))).((((--(......)))))........ ( -25.40) >DroSim_CAF1 23216 93 - 1 ACAGUUCUUGC-------------------------AGCUUAAUGCUUAAUGGCACGCCACGUGCAACGUAGCAUUAAAGUGGCAAUCAAAGCGGUUG--AAACACAUCAACACUUCAUC ...(((.((((-------------------------.((((((((((.....(((((...))))).....)))))).)))).))))....))).((((--(......)))))........ ( -25.40) >DroEre_CAF1 38592 92 - 1 AC-GUUCUUGC-------------------------AGCUUAAUGCUUAAUAGCACGCUACGUGCAACGUUGCAUUAAAGUGGCAAUCAAAGUGGUUG--AAUCACAUCAACACUUCAUC ..-....((((-------------------------.(((((((((......(((((...)))))......))))).)))).))))...(((((.(((--(......))))))))).... ( -24.30) >DroYak_CAF1 30700 93 - 1 ACAGUUCUUGC-------------------------AGCUUAAUGCUUAAUAGCACGCUACGUGCAACGUAGCAUUAAAGUGGCAAUCAAAGUGGUUG--AAUCACAUCAACACUUCAUC .......((((-------------------------.((((((((((.....(((((...))))).....)))))).)))).))))...(((((.(((--(......))))))))).... ( -25.90) >DroPer_CAF1 43760 120 - 1 CCAAUUUUUGCGGAGCAGCAUCAGCAGCAGCAAAGGAGCUUAAUGCAUAAUAACAUGCCACGUGCAACAUCGCAUUAAAAUGCCAAUCAAAGUGGUUGUGGAAACCAUCAGCACUUCAUC ....(((((((...((.((....)).)).)))))))........((((......))))...((((......((((....))))........((((((.....))))))..))))...... ( -25.80) >consensus ACAGUUCUUGC_________________________AGCUUAAUGCUUAAUAGCACGCCACGUGCAACGUAGCAUUAAAGUGGCAAUCAAAGCGGUUG__AAACACAUCAACACUUCAUC ...(((..((.............................((((((((.....(((((...))))).....)))))))).....(((((.....))))).......))..)))........ (-15.31 = -15.57 + 0.25)

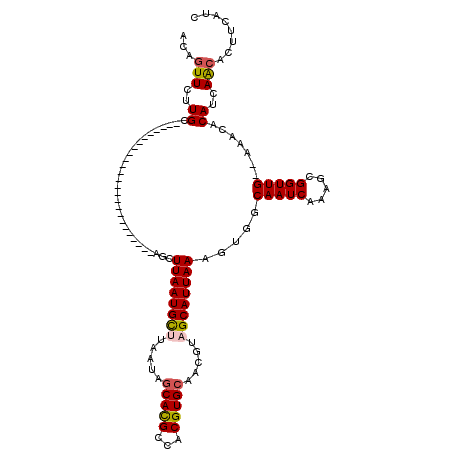
| Location | 22,179,893 – 22,179,988 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 89.86 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22179893 95 + 27905053 CUUUAAUGCUACGUUGCACGUAGCGUGCCAUUAAACAUUAAGCUGCAAGAACGGUAGCACAAAAAACUAUAGGCUUACGGAUUUUAUCGAGAGUU .(((((((.(((((((....))))))).)))))))......(((((.......))))).............(((((.(((......))).))))) ( -21.60) >DroSec_CAF1 24407 95 + 1 CUUUAAUGCUACGUUGCACGUGGCGUGCCAUUAAGCAUUAAGCUGCAAGAACUGUAGCAUAAAAAACUGUAGGCUUACGGAUUUUAUCGUGAGUA ..((((((((..((.(((((...))))).))..))))))))((((((.....))))))..............((((((((......)))))))). ( -29.50) >DroSim_CAF1 23254 95 + 1 CUUUAAUGCUACGUUGCACGUGGCGUGCCAUUAAGCAUUAAGCUGCAAGAACUGUAGCAUAAAAAACUGUAGGCUUACGGCUUGUAUCGUGAGUU ..((((((((..((.(((((...))))).))..))))))))((((((.....)))))).............(((((((((......))))))))) ( -29.50) >DroEre_CAF1 38630 89 + 1 CUUUAAUGCAACGUUGCACGUAGCGUGCUAUUAAGCAUUAAGCUGCAAGAAC-GUAGCACAAAAAA-----GGCUCACGGUUUUUAUCGAGAGUU ..(((((((......(((((...)))))......)))))))(((((......-)))))........-----(((((.((((....)))).))))) ( -27.00) >DroYak_CAF1 30738 92 + 1 CUUUAAUGCUACGUUGCACGUAGCGUGCUAUUAAGCAUUAAGCUGCAAGAACUGUAGCACAAAAAAAC---GGCUUACGGAUUUUAUCAAGAGUU ..((((((((.....(((((...))))).....))))))))((((((.....))))))...((((..(---(.....))..)))).......... ( -24.40) >consensus CUUUAAUGCUACGUUGCACGUAGCGUGCCAUUAAGCAUUAAGCUGCAAGAACUGUAGCACAAAAAACU_UAGGCUUACGGAUUUUAUCGAGAGUU ..((((((((.....(((((...))))).....))))))))(((((.......)))))..............((((.(((......))).)))). (-21.46 = -21.90 + 0.44)

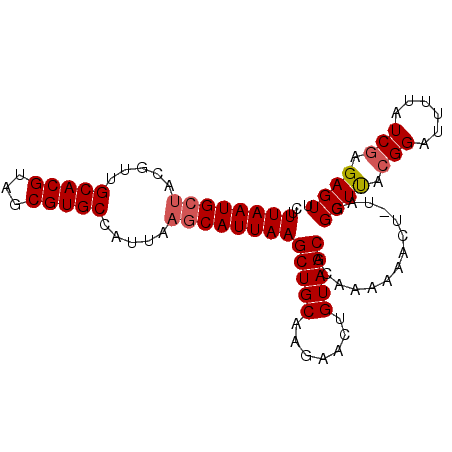
| Location | 22,179,893 – 22,179,988 |
|---|---|
| Length | 95 |
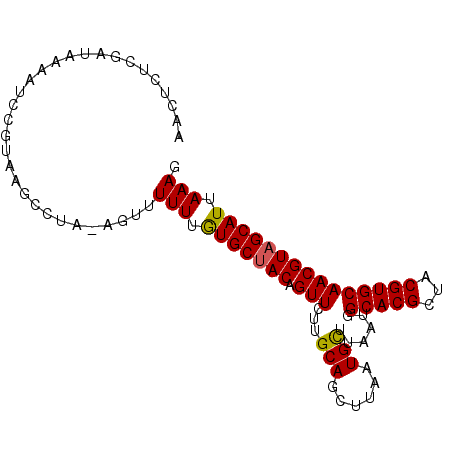
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 89.86 |
| Mean single sequence MFE | -21.06 |
| Consensus MFE | -17.58 |
| Energy contribution | -17.38 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22179893 95 - 27905053 AACUCUCGAUAAAAUCCGUAAGCCUAUAGUUUUUUGUGCUACCGUUCUUGCAGCUUAAUGUUUAAUGGCACGCUACGUGCAACGUAGCAUUAAAG ..((..((........))..)).........(((.(((((((((((...(((......)))..))))(((((...)))))...))))))).))). ( -18.90) >DroSec_CAF1 24407 95 - 1 UACUCACGAUAAAAUCCGUAAGCCUACAGUUUUUUAUGCUACAGUUCUUGCAGCUUAAUGCUUAAUGGCACGCCACGUGCAACGUAGCAUUAAAG ..((.(((........))).)).........(((.(((((((.......(((......)))......(((((...)))))...))))))).))). ( -20.40) >DroSim_CAF1 23254 95 - 1 AACUCACGAUACAAGCCGUAAGCCUACAGUUUUUUAUGCUACAGUUCUUGCAGCUUAAUGCUUAAUGGCACGCCACGUGCAACGUAGCAUUAAAG ..............((.(((((.((..(((.......)))..))..))))).))((((((((.....(((((...))))).....)))))))).. ( -22.70) >DroEre_CAF1 38630 89 - 1 AACUCUCGAUAAAAACCGUGAGCC-----UUUUUUGUGCUAC-GUUCUUGCAGCUUAAUGCUUAAUAGCACGCUACGUGCAACGUUGCAUUAAAG ..(((.((........)).))).(-----(((...((((.((-(((...(((......)))......(((((...)))))))))).)))).)))) ( -23.00) >DroYak_CAF1 30738 92 - 1 AACUCUUGAUAAAAUCCGUAAGCC---GUUUUUUUGUGCUACAGUUCUUGCAGCUUAAUGCUUAAUAGCACGCUACGUGCAACGUAGCAUUAAAG .....(((((((((..((.....)---)..)))).((((((..((....))(((.....)))...))))))((((((.....))))))))))).. ( -20.30) >consensus AACUCUCGAUAAAAUCCGUAAGCCUA_AGUUUUUUGUGCUACAGUUCUUGCAGCUUAAUGCUUAAUGGCACGCUACGUGCAACGUAGCAUUAAAG ...............................(((.(((((((.(((...(((......)))......(((((...))))))))))))))).))). (-17.58 = -17.38 + -0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:25 2006