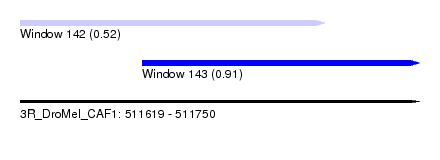
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 511,619 – 511,750 |
| Length | 131 |
| Max. P | 0.910471 |

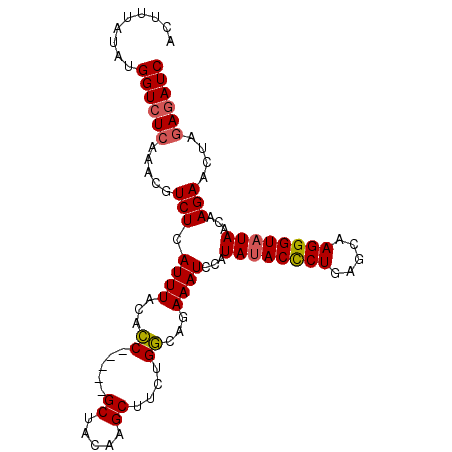
| Location | 511,619 – 511,719 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 84.41 |
| Mean single sequence MFE | -19.00 |
| Consensus MFE | -13.16 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 511619 100 + 27905053 ACUUUAUAUGGUAUCAAACGUCUCAUUUACUGCUAUAAGCUACAAGCUUCUGUCGGAAAUCCAUAAACCCUGAGCAAGGGUAUAACAAGAACUAUAGAUC ......((((((.((..............(((.((.((((.....)))).)).)))..........(((((.....))))).......)))))))).... ( -18.10) >DroSec_CAF1 38314 95 + 1 ACUUUAUAUGGUCUCAAACUUCUCAUUUACACC-----GCUACAAGCUUCUGGCAAAAAUCCAUAUACUCUAAGCAAGGGUAUAACAAGAACUAGAGAUC .........((((((....((((..........-----((((........)))).........((((((((.....))))))))...))))...)))))) ( -17.30) >DroSim_CAF1 38345 88 + 1 -------AUGGUCUCAAACGUCUCAUUUACACC-----GCUACAAGCUUCUGGCAGAAAUCCAUAUACCCUGAGCAAGGGUAUAACAAGAACUAGAGAUC -------..((((((..................-----((.....))((((((.(....))).((((((((.....))))))))...))))...)))))) ( -21.60) >consensus ACUUUAUAUGGUCUCAAACGUCUCAUUUACACC_____GCUACAAGCUUCUGGCAGAAAUCCAUAUACCCUGAGCAAGGGUAUAACAAGAACUAGAGAUC .........((((((.....(((.((((...((.....((.....))....))...))))...((((((((.....))))))))...)))....)))))) (-13.16 = -13.50 + 0.34)

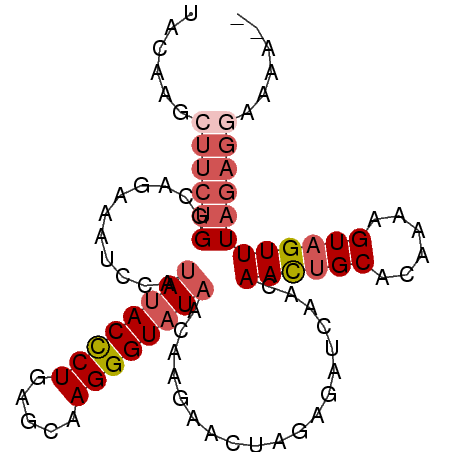
| Location | 511,659 – 511,750 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -18.50 |
| Consensus MFE | -12.52 |
| Energy contribution | -14.40 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 511659 91 + 27905053 UACAAGCUUCUGUCGGAAAUCCAUAAACCCUGAGCAAGGGUAUAACAAGAACUAUAGAUCAAUAACUGCACAAAAGUAGUUUAGAGUAAAA-- (((.....(((((.((....))....(((((.....)))))............)))))((...((((((......))))))..)))))...-- ( -18.00) >DroSec_CAF1 38349 93 + 1 UACAAGCUUCUGGCAAAAAUCCAUAUACUCUAAGCAAGGGUAUAACAAGAACUAGAGAUCAACAACUGCACAAAAGUAGUUUAGAGGAAAAUU ......((((((...........((((((((.....))))))))...................((((((......))))))))))))...... ( -17.20) >DroSim_CAF1 38373 93 + 1 UACAAGCUUCUGGCAGAAAUCCAUAUACCCUGAGCAAGGGUAUAACAAGAACUAGAGAUCAACAACUGCACAAAAGUAGUUUAGAGGAAAAUU ......((((((...........((((((((.....))))))))...................((((((......))))))))))))...... ( -21.10) >DroEre_CAF1 42485 86 + 1 UACAAACUUCUGACUGAAUUCUAUAUACCCUGAGCAAGGGUAUUAGAAGUACUAGAU----ACAAUGGCACAAAAGUAGUUUUCCUCUAC--- ...(((((.((.......(((((.(((((((.....))))))))))))((.(((...----....))).))...)).)))))........--- ( -17.70) >consensus UACAAGCUUCUGGCAGAAAUCCAUAUACCCUGAGCAAGGGUAUAACAAGAACUAGAGAUCAACAACUGCACAAAAGUAGUUUAGAGGAAAA__ ......((((((...........((((((((.....))))))))...................((((((......))))))))))))...... (-12.52 = -14.40 + 1.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:37 2006