
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,151,656 – 22,151,854 |
| Length | 198 |
| Max. P | 0.919488 |

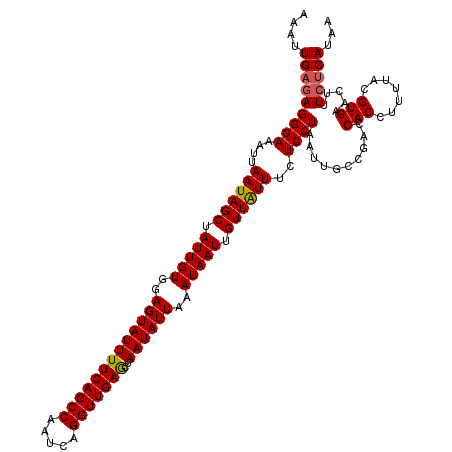
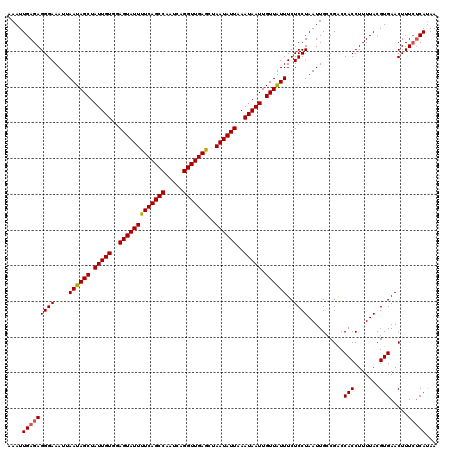
| Location | 22,151,656 – 22,151,774 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -26.04 |
| Energy contribution | -26.27 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22151656 118 - 27905053 AAAUUGAGAGGGAAAUUAAUAGCUAUUGUGGAGUAUUUUCAGCCAAUCAGGUUGAACUAAUAUUAAAUAAUUGUUGUUUCUCCUAAUUGGCGGCCACCUUUUACGUGAACUUU--CAAAA ...((((((((((....((((((.(((((..(((((((((((((.....)))))))..))))))..))))).))))))..)))...((.(((...........))).))))))--))).. ( -30.90) >DroSec_CAF1 82105 120 - 1 AAAUUGAGAGGGAAAUUAAUAGCUAUUGUGGAGUAUUUUCAGCCAAUCAGGUUGAGCUAAUAUUAAAUAAUUGUUAUUUCUCCUAAUUGCCGACCACCUUUUACGUGAACUUUCUCAUAA ....(((((((((....((((((.(((((..(((((((((((((.....)))))))..))))))..))))).))))))..))))..........(((.......))).....)))))... ( -28.40) >DroSim_CAF1 76588 120 - 1 AAAUUGAGAGGGAAAUUAAUAGCUAUUGUGGAGUAUUUUCAGCCAAUCAGGUUGAGCUAAUAUUAAAUAAUUGUUAUUUCUCCUAAUUGCCGACCACCUUUUACGUGAACUUUCUCAUAA ....(((((((((....((((((.(((((..(((((((((((((.....)))))))..))))))..))))).))))))..))))..........(((.......))).....)))))... ( -28.40) >consensus AAAUUGAGAGGGAAAUUAAUAGCUAUUGUGGAGUAUUUUCAGCCAAUCAGGUUGAGCUAAUAUUAAAUAAUUGUUAUUUCUCCUAAUUGCCGACCACCUUUUACGUGAACUUUCUCAUAA ....(((((((((....((((((.(((((..(((((((((((((.....)))))))..))))))..))))).))))))..))))..........(((.......))).....)))))... (-26.04 = -26.27 + 0.23)

| Location | 22,151,694 – 22,151,814 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -27.94 |
| Energy contribution | -27.50 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.95 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22151694 120 - 27905053 UACAAAUAGUUCUAAGCUGAUUGCUCUCUAAUUACAGUGCAAAUUGAGAGGGAAAUUAAUAGCUAUUGUGGAGUAUUUUCAGCCAAUCAGGUUGAACUAAUAUUAAAUAAUUGUUGUUUC ..(((.((((.....)))).)))((((((.....((((....)))))))))).....((((((.(((((..(((((((((((((.....)))))))..))))))..))))).)))))).. ( -27.70) >DroSec_CAF1 82145 120 - 1 UACAAAUAGUUCUAAGCUGAUUGCUCUCUAAAUUCAGUGCAAAUUGAGAGGGAAAUUAAUAGCUAUUGUGGAGUAUUUUCAGCCAAUCAGGUUGAGCUAAUAUUAAAUAAUUGUUAUUUC ..(((.((((.....)))).)))(((((.....(((((....)))))))))).....((((((.(((((..(((((((((((((.....)))))))..))))))..))))).)))))).. ( -27.70) >DroSim_CAF1 76628 120 - 1 UACAAAUAGUUCUAAGCUGAUUGCUCUCUAAUUACAGUGCAAAUUGAGAGGGAAAUUAAUAGCUAUUGUGGAGUAUUUUCAGCCAAUCAGGUUGAGCUAAUAUUAAAUAAUUGUUAUUUC ..(((.((((.....)))).)))((((((.....((((....)))))))))).....((((((.(((((..(((((((((((((.....)))))))..))))))..))))).)))))).. ( -27.40) >consensus UACAAAUAGUUCUAAGCUGAUUGCUCUCUAAUUACAGUGCAAAUUGAGAGGGAAAUUAAUAGCUAUUGUGGAGUAUUUUCAGCCAAUCAGGUUGAGCUAAUAUUAAAUAAUUGUUAUUUC ..(((.((((.....)))).)))((((((.....((((....)))))))))).....((((((.(((((..(((((((((((((.....)))))))..))))))..))))).)))))).. (-27.94 = -27.50 + -0.44)

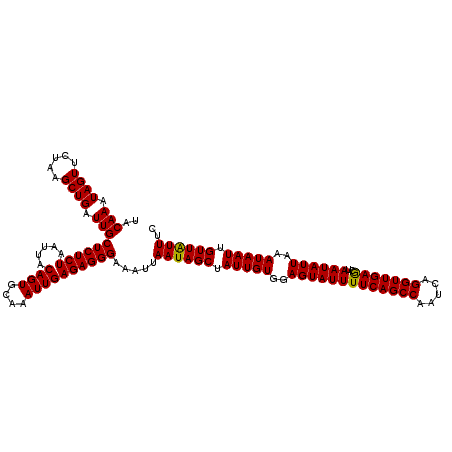
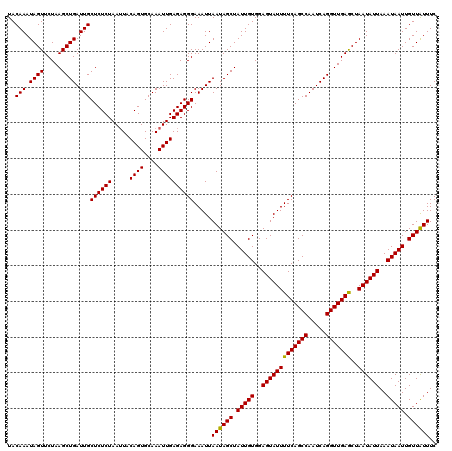
| Location | 22,151,734 – 22,151,854 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -24.11 |
| Consensus MFE | -23.43 |
| Energy contribution | -23.77 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22151734 120 - 27905053 UCAACCUCAACAGCUUCAUAUGUAUUUCCAUUAACAAGUCUACAAAUAGUUCUAAGCUGAUUGCUCUCUAAUUACAGUGCAAAUUGAGAGGGAAAUUAAUAGCUAUUGUGGAGUAUUUUC ....((.(((.((((........((((((.............(((.((((.....)))).)))(((((.((((........)))))))))))))))....)))).))).))......... ( -24.50) >DroSec_CAF1 82185 120 - 1 UCAACCUCAACAGCUUCAUAUGUAUUUCCAUUAACAACUCUACAAAUAGUUCUAAGCUGAUUGCUCUCUAAAUUCAGUGCAAAUUGAGAGGGAAAUUAAUAGCUAUUGUGGAGUAUUUUC ....((.(((.((((........((((((.............(((.((((.....)))).)))(((((.................)))))))))))....)))).))).))......... ( -23.33) >DroSim_CAF1 76668 120 - 1 UCAACCUCAACAGCUUCAUAUGUAUUUCCAUUAACAACUCUACAAAUAGUUCUAAGCUGAUUGCUCUCUAAUUACAGUGCAAAUUGAGAGGGAAAUUAAUAGCUAUUGUGGAGUAUUUUC ....((.(((.((((........((((((.............(((.((((.....)))).)))(((((.((((........)))))))))))))))....)))).))).))......... ( -24.50) >consensus UCAACCUCAACAGCUUCAUAUGUAUUUCCAUUAACAACUCUACAAAUAGUUCUAAGCUGAUUGCUCUCUAAUUACAGUGCAAAUUGAGAGGGAAAUUAAUAGCUAUUGUGGAGUAUUUUC ....((.(((.((((........((((((.............(((.((((.....)))).)))(((((.((((........)))))))))))))))....)))).))).))......... (-23.43 = -23.77 + 0.33)

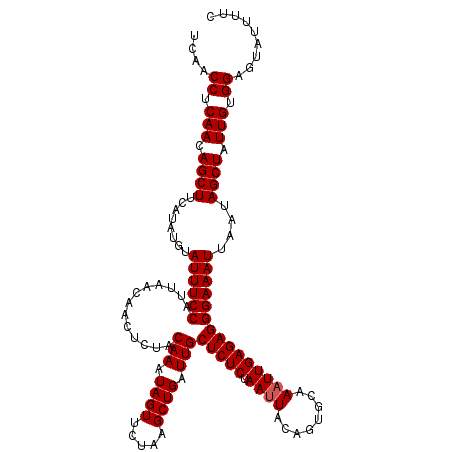
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:13 2006