
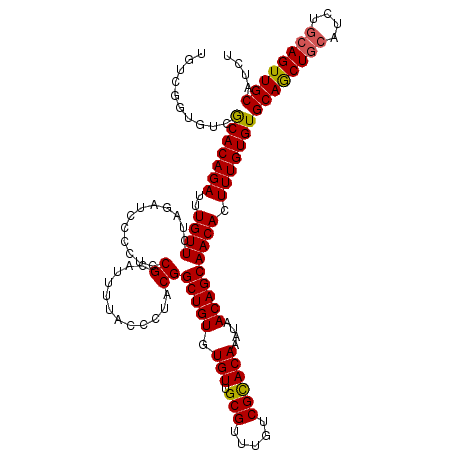
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,149,766 – 22,149,925 |
| Length | 159 |
| Max. P | 0.995425 |

| Location | 22,149,766 – 22,149,885 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.39 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -31.06 |
| Energy contribution | -31.00 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22149766 119 + 27905053 UGUCGGGGUUGCACAGAUUUGUUCUAGA-CCCCUCCGCAUUUUACCCUACGGCUGUGUGUUGCGUUUGUCGCACAAAUAACAGCAACACUUUGUGUGCAACUGCAUCUGCAGUUGCAUCU ....((((((....(((.....))).))-))))..................(((((.(((.(((.....))))))....)))))..........((((((((((....)))))))))).. ( -40.30) >DroSec_CAF1 80215 120 + 1 UGUCGGUGUCGCACAGAUUUGUUCUAGCUCCCCUCCGCAUUUUACCCUACGGCUGUGUGUUGCGUUUGUCGCACAAAUAACAGCAACACUUUGUGUGCAGCUGCAUCUGCAGUUGCAUCU ....(((...(((((((..((((...((........)).............(((((.(((.(((.....))))))....))))))))).)))))))((((((((....))))))))))). ( -36.90) >DroSim_CAF1 74690 114 + 1 UGUCGGUGUCGCACAGAUUUGUUCUAGCUCCCCUUCGCAUUUUACCCUACGGCUGUGUGUUGCGUUUGUCGCACAAAUAACAGCAACACUUUGUGUGCAGCU------GCAGUUGCAUCU (((.(.((.((((((((..((((...((........)).............(((((.(((.(((.....))))))....))))))))).)))))))))).).------)))......... ( -29.90) >DroYak_CAF1 73136 119 + 1 UGUCGGUGUCACACAGAUUUGUUCUAGACUCCCUGCGCAUUU-CCCCUACGGCUGUGUGUUGCGUUUGCCGUACAUAUAACAGCAACACUUUGUGUGCAGCUGCAUCUGCAGUUGCAUCU .(((((((...))).((..(((..(((.....))).)))..)-).....))))...((((((((((............))).))))))).....((((((((((....)))))))))).. ( -33.70) >consensus UGUCGGUGUCGCACAGAUUUGUUCUAGAUCCCCUCCGCAUUUUACCCUACGGCUGUGUGUUGCGUUUGUCGCACAAAUAACAGCAACACUUUGUGUGCAGCUGCAUCUGCAGUUGCAUCU ..........(((((((..((((............((............))(((((.(((.(((.....))))))....))))))))).)))))))((((((((....)))))))).... (-31.06 = -31.00 + -0.06)

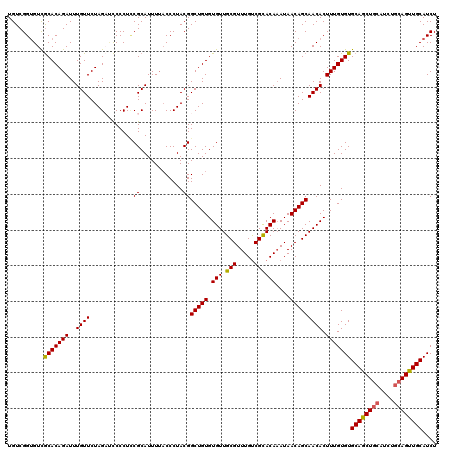
| Location | 22,149,766 – 22,149,885 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.39 |
| Mean single sequence MFE | -35.34 |
| Consensus MFE | -26.24 |
| Energy contribution | -27.55 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22149766 119 - 27905053 AGAUGCAACUGCAGAUGCAGUUGCACACAAAGUGUUGCUGUUAUUUGUGCGACAAACGCAACACACAGCCGUAGGGUAAAAUGCGGAGGGG-UCUAGAACAAAUCUGUGCAACCCCGACA ...(((((((((....)))))))))......(((((((((((........))))...)))))))....(((((........))))).((((-(.....(((....)))...))))).... ( -41.80) >DroSec_CAF1 80215 120 - 1 AGAUGCAACUGCAGAUGCAGCUGCACACAAAGUGUUGCUGUUAUUUGUGCGACAAACGCAACACACAGCCGUAGGGUAAAAUGCGGAGGGGAGCUAGAACAAAUCUGUGCGACACCGACA ...((((.((((....)))).))))......(((((((((((........))))...)))))))....(((((........))))).(((..((((((.....)))).))..).)).... ( -36.30) >DroSim_CAF1 74690 114 - 1 AGAUGCAACUGC------AGCUGCACACAAAGUGUUGCUGUUAUUUGUGCGACAAACGCAACACACAGCCGUAGGGUAAAAUGCGAAGGGGAGCUAGAACAAAUCUGUGCGACACCGACA .(.((((...((------(((.((((.....)))).)))))......)))).)...((((.......(((....)))....))))..(((..((((((.....)))).))..).)).... ( -30.00) >DroYak_CAF1 73136 119 - 1 AGAUGCAACUGCAGAUGCAGCUGCACACAAAGUGUUGCUGUUAUAUGUACGGCAAACGCAACACACAGCCGUAGGGG-AAAUGCGCAGGGAGUCUAGAACAAAUCUGUGUGACACCGACA ...((((.((((....)))).)))).......(((((.((((...(.((((((..............)))))).)..-....(((((((..((.....))...))))))))))).))))) ( -33.24) >consensus AGAUGCAACUGCAGAUGCAGCUGCACACAAAGUGUUGCUGUUAUUUGUGCGACAAACGCAACACACAGCCGUAGGGUAAAAUGCGGAGGGGAGCUAGAACAAAUCUGUGCGACACCGACA ...((((.((((....)))).))))......(((((((((((........))))...)))))))....(((((........))))).(((..((((((.....)))).))..).)).... (-26.24 = -27.55 + 1.31)

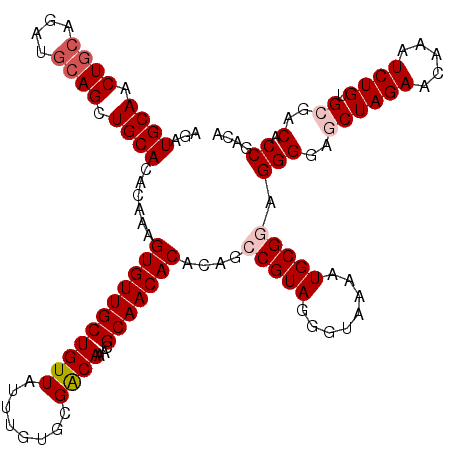

| Location | 22,149,805 – 22,149,925 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -38.06 |
| Energy contribution | -38.62 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22149805 120 + 27905053 UUUACCCUACGGCUGUGUGUUGCGUUUGUCGCACAAAUAACAGCAACACUUUGUGUGCAACUGCAUCUGCAGUUGCAUCUGGCAUGUUAUUUGCUGUCUGAUGAUACCUAAACCCAGCCC ..........(((((.((...(.(((..((((((((((((((((.((((...))))((((((((....)))))))).....)).))))))))).)))..))..).)))...)).))))). ( -41.90) >DroSec_CAF1 80255 120 + 1 UUUACCCUACGGCUGUGUGUUGCGUUUGUCGCACAAAUAACAGCAACACUUUGUGUGCAGCUGCAUCUGCAGUUGCAUCUGGCAUGUUAUUUGCUGUCUGAUGAUACCUAAACCCAGCCC ..........(((((.((...(.(((..((((((((((((((((.((((...))))((((((((....)))))))).....)).))))))))).)))..))..).)))...)).))))). ( -41.90) >DroSim_CAF1 74730 114 + 1 UUUACCCUACGGCUGUGUGUUGCGUUUGUCGCACAAAUAACAGCAACACUUUGUGUGCAGCU------GCAGUUGCAUCUGGCAUGUUAUUUGCUGUCUGAUGAUACCUAAACCCAGCCC ..........(((((.((...(.(((..((((((((((((((((..(((.....)))(((.(------((....))).))))).))))))))).)))..))..).)))...)).))))). ( -34.40) >DroYak_CAF1 73176 119 + 1 UU-CCCCUACGGCUGUGUGUUGCGUUUGCCGUACAUAUAACAGCAACACUUUGUGUGCAGCUGCAUCUGCAGUUGCAUCUGGCAUGUUAUUUGCUGUCUGAUGAUACCUAAACCCAGCCC ..-.......(((((.((((((((((............))).)))))))...((((((((((((....))))))))(((.((((.((.....)))))).))).)))).......))))). ( -39.40) >consensus UUUACCCUACGGCUGUGUGUUGCGUUUGUCGCACAAAUAACAGCAACACUUUGUGUGCAGCUGCAUCUGCAGUUGCAUCUGGCAUGUUAUUUGCUGUCUGAUGAUACCUAAACCCAGCCC ..........(((((.((((((((((((.....)))))....)))))))...((((((((((((....))))))))(((.((((.((.....)))))).))).)))).......))))). (-38.06 = -38.62 + 0.56)



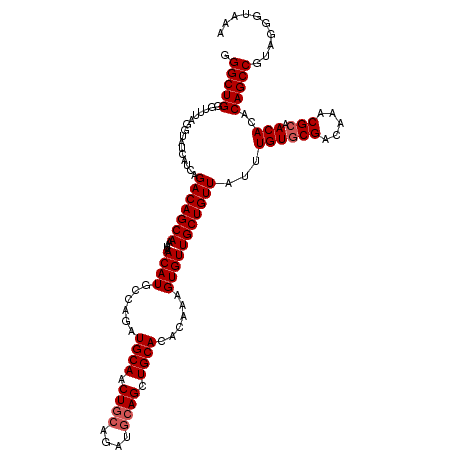
| Location | 22,149,805 – 22,149,925 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -32.40 |
| Energy contribution | -33.15 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22149805 120 - 27905053 GGGCUGGGUUUAGGUAUCAUCAGACAGCAAAUAACAUGCCAGAUGCAACUGCAGAUGCAGUUGCACACAAAGUGUUGCUGUUAUUUGUGCGACAAACGCAACACACAGCCGUAGGGUAAA .(((((.(((................((((((((((.((.....((((((((....))))))))((((...)))).))))))))))))(((.....))))))...))))).......... ( -40.70) >DroSec_CAF1 80255 120 - 1 GGGCUGGGUUUAGGUAUCAUCAGACAGCAAAUAACAUGCCAGAUGCAACUGCAGAUGCAGCUGCACACAAAGUGUUGCUGUUAUUUGUGCGACAAACGCAACACACAGCCGUAGGGUAAA .(((((.(((................((((((((((.((.....(((.((((....)))).)))((((...)))).))))))))))))(((.....))))))...))))).......... ( -36.50) >DroSim_CAF1 74730 114 - 1 GGGCUGGGUUUAGGUAUCAUCAGACAGCAAAUAACAUGCCAGAUGCAACUGC------AGCUGCACACAAAGUGUUGCUGUUAUUUGUGCGACAAACGCAACACACAGCCGUAGGGUAAA .(((((.(((................((((((((((.(((((.(((....))------).)))(((.....)))..))))))))))))(((.....))))))...))))).......... ( -32.40) >DroYak_CAF1 73176 119 - 1 GGGCUGGGUUUAGGUAUCAUCAGACAGCAAAUAACAUGCCAGAUGCAACUGCAGAUGCAGCUGCACACAAAGUGUUGCUGUUAUAUGUACGGCAAACGCAACACACAGCCGUAGGGG-AA .(((((.(((...(((.(((..(((((((....((((......((((.((((....)))).))))......)))))))))))..)))))).((....)))))...))))).......-.. ( -36.80) >consensus GGGCUGGGUUUAGGUAUCAUCAGACAGCAAAUAACAUGCCAGAUGCAACUGCAGAUGCAGCUGCACACAAAGUGUUGCUGUUAUUUGUGCGACAAACGCAACACACAGCCGUAGGGUAAA .(((((................(((((((....((((......((((.((((....)))).))))......)))))))))))...((((((.....))).)))..))))).......... (-32.40 = -33.15 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:08 2006