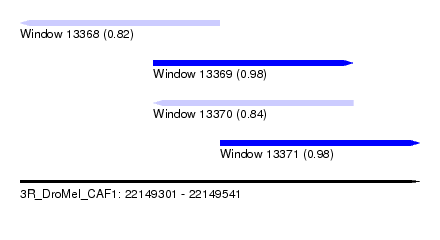
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,149,301 – 22,149,541 |
| Length | 240 |
| Max. P | 0.977558 |

| Location | 22,149,301 – 22,149,421 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -27.74 |
| Energy contribution | -28.80 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22149301 120 - 27905053 GGCAAAUUAUACCUUCUUCAAAGCUUUAAAGACCUUUUAAGGCACUUCGGCCCCAGGGGAGGAGGGCAAGAAAGUUGUUCAAUCGAAACGACAAUUAAUUGGCUAAAAUAAAUAUUCACG (((.(((((..((((((((...(((((((((...))))))))).......((....)))))))))).......(((((((....).))))))...))))).)))................ ( -28.90) >DroSec_CAF1 79752 120 - 1 GGCAAAUUAUGACCUCUUCAGAGCUUUAAAGACCUUUUAAGGCACUUCGGCCCCAGGGGAGGAGGGCAAGAAAGUUGUUCAAUAGAAACGACAAUUAAUUGGCUAAAAUAAAUAUUCACG (((.(((((((.(((((((...(((((((((...))))))))).......((....))))))))).)).....(((((((....).))))))...))))).)))................ ( -29.70) >DroSim_CAF1 74227 120 - 1 GGCAAAUUAUGCCCUCUUCAGAGCUUUAAAGACCUUUUAAGUCACUUCGGCCCCAGGGGAGGAGGGCAAGAAAGUUGUUCAAUCGAAACGACAAUUAAUUGGCUAAAAUAAAUAUUCACG (((.(((((((((((((((...........(((.......))).......((....)))))))))))).....(((((((....).))))))...))))).)))................ ( -32.00) >DroYak_CAF1 72666 120 - 1 GGCAAAUUAUGGCCUCUUCAAAGCUUUAAAGGCCUUUUAAGGCUCUUCGCCCCCAGGGGAGAAGGGCAAGAAAGUUGUUCAAUCGAAACGACAAUUAAUUGGCUAAAAUAAAUAUUCACG (((........)))................((((..((((.(((((((.(((....))).)))))))......(((((((....).))))))..))))..))))................ ( -34.50) >consensus GGCAAAUUAUGCCCUCUUCAAAGCUUUAAAGACCUUUUAAGGCACUUCGGCCCCAGGGGAGGAGGGCAAGAAAGUUGUUCAAUCGAAACGACAAUUAAUUGGCUAAAAUAAAUAUUCACG (((.(((((((((((((((...(((((((((...))))))))).......((....)))))))))))).....(((((((....).))))))...))))).)))................ (-27.74 = -28.80 + 1.06)

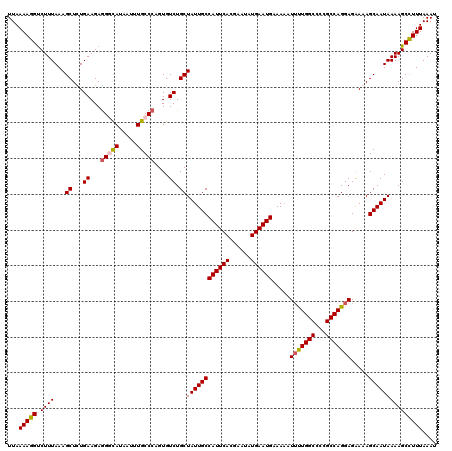
| Location | 22,149,381 – 22,149,501 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -28.84 |
| Energy contribution | -29.28 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22149381 120 + 27905053 UUAAAAGGUCUUUAAAGCUUUGAAGAAGGUAUAAUUUGCCCUGUGUCUGCUAUUGCCAUUCACGAAUAUGAAUGAAAAAUUUUGGCCCCGCCAGGAGAAAAGCAAUAAAAGCCUUUAAAU ...(((((.((((...((...((....((((.....)))).....)).))((((((((((((......)))))).....(((((((...))))))).....))))))))))))))).... ( -29.20) >DroSec_CAF1 79832 120 + 1 UUAAAAGGUCUUUAAAGCUCUGAAGAGGUCAUAAUUUGCCCAGUGUCUGCUAUUGCCAUUCACGAAUAUGAAUGAAAAAUGUUGGCCCCGCCAGGAGAAAAGCAAUAAAAGCCUUUAAAU ...(((((.((((...((...((...((.((.....)).))....)).))((((((((((((......)))))).....(.(((((...))))).).....))))))))))))))).... ( -26.20) >DroSim_CAF1 74307 120 + 1 UUAAAAGGUCUUUAAAGCUCUGAAGAGGGCAUAAUUUGCCCAGUGUCUGCUAUUGCCAUUCACGAAUAUGAAUGAAAAAUUCUGGCCCCGCCAGGAGAAAAGCAAUAAAAGCCUUUAAAU ...(((((.((((...((...((...(((((.....)))))....)).))((((((((((((......)))))).....(((((((...))))))).....))))))))))))))).... ( -37.20) >DroYak_CAF1 72746 120 + 1 UUAAAAGGCCUUUAAAGCUUUGAAGAGGCCAUAAUUUGCCCAGUGUCUGCUAUUGCCAUUCACGAAUAUGAAUGAAAUUUUUUGGCCCCGCCAGGAGGAAAGCAAUAAAAGCUUUUAAAU (((((((((((((..........)))))))..............(.((..((((((((((((......))))))...(((((((((...)))))))))...))))))..))))))))).. ( -29.30) >consensus UUAAAAGGUCUUUAAAGCUCUGAAGAGGGCAUAAUUUGCCCAGUGUCUGCUAUUGCCAUUCACGAAUAUGAAUGAAAAAUUUUGGCCCCGCCAGGAGAAAAGCAAUAAAAGCCUUUAAAU ...(((((.((((...((...((...(((((.....)))))....)).))((((((((((((......)))))).....(((((((...))))))).....))))))))))))))).... (-28.84 = -29.28 + 0.44)

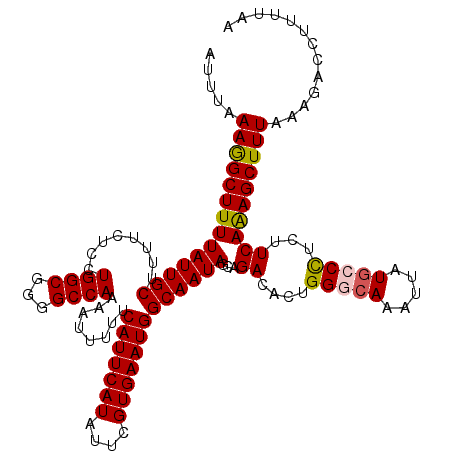
| Location | 22,149,381 – 22,149,501 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -25.23 |
| Energy contribution | -25.35 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22149381 120 - 27905053 AUUUAAAGGCUUUUAUUGCUUUUCUCCUGGCGGGGCCAAAAUUUUUCAUUCAUAUUCGUGAAUGGCAAUAGCAGACACAGGGCAAAUUAUACCUUCUUCAAAGCUUUAAAGACCUUUUAA ....(((((..((((..(((((..(((....)))(((.........(((((((....)))))))((....))........)))................)))))..))))..)))))... ( -25.80) >DroSec_CAF1 79832 120 - 1 AUUUAAAGGCUUUUAUUGCUUUUCUCCUGGCGGGGCCAACAUUUUUCAUUCAUAUUCGUGAAUGGCAAUAGCAGACACUGGGCAAAUUAUGACCUCUUCAGAGCUUUAAAGACCUUUUAA ....(((((..((((..(((......((((.((((.((........(((((((....)))))))((..(((......))).))......)).)))).)))))))..))))..)))))... ( -26.90) >DroSim_CAF1 74307 120 - 1 AUUUAAAGGCUUUUAUUGCUUUUCUCCUGGCGGGGCCAGAAUUUUUCAUUCAUAUUCGUGAAUGGCAAUAGCAGACACUGGGCAAAUUAUGCCCUCUUCAGAGCUUUAAAGACCUUUUAA .....((((((((((((((.......(((((...))))).......(((((((....)))))))))))))...((....(((((.....)))))...))))))))))............. ( -33.80) >DroYak_CAF1 72746 120 - 1 AUUUAAAAGCUUUUAUUGCUUUCCUCCUGGCGGGGCCAAAAAAUUUCAUUCAUAUUCGUGAAUGGCAAUAGCAGACACUGGGCAAAUUAUGGCCUCUUCAAAGCUUUAAAGGCCUUUUAA ..(((((((((((((..(((((((....)).(((((((........(((((((....)))))))((..(((......))).))......)))))))...)))))..)))))).))))))) ( -35.50) >consensus AUUUAAAGGCUUUUAUUGCUUUUCUCCUGGCGGGGCCAAAAUUUUUCAUUCAUAUUCGUGAAUGGCAAUAGCAGACACUGGGCAAAUUAUGCCCUCUUCAAAGCUUUAAAGACCUUUUAA .....((((((((((((((........((((...))))........(((((((....)))))))))))))...((....(((((.....)))))...))))))))))............. (-25.23 = -25.35 + 0.12)

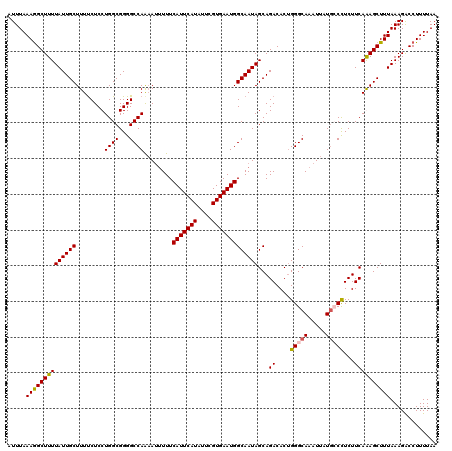
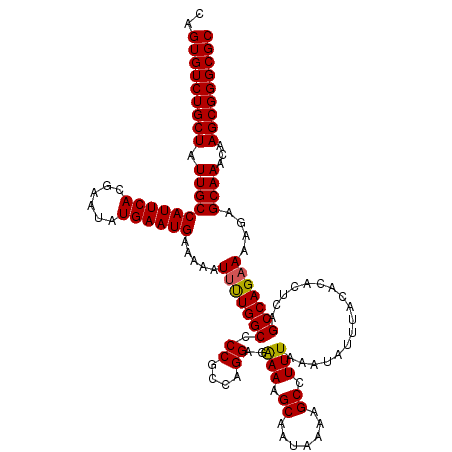
| Location | 22,149,421 – 22,149,541 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.08 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -30.95 |
| Energy contribution | -30.82 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22149421 120 + 27905053 CUGUGUCUGCUAUUGCCAUUCACGAAUAUGAAUGAAAAAUUUUGGCCCCGCCAGGAGAAAAGCAAUAAAAGCCUUUAAAUAUUUACACACUCAGCCAGAAAAGAGCAAACAAGCGGGCGC ..(((((((((.((((((((((......)))))).....(((((((.((....))..(((.((.......)).))).................)))))))....))))...))))))))) ( -30.50) >DroSec_CAF1 79872 120 + 1 CAGUGUCUGCUAUUGCCAUUCACGAAUAUGAAUGAAAAAUGUUGGCCCCGCCAGGAGAAAAGCAAUAAAAGCCUUUAAAUAUUUACACACUCAGCCAGAAAAGAGCAAACAAGCGGGCGC ..((((((((((((((((((((......)))))).....(.(((((...))))).).....))))))...((((((.......................)))).))......)))))))) ( -28.60) >DroSim_CAF1 74347 120 + 1 CAGUGUCUGCUAUUGCCAUUCACGAAUAUGAAUGAAAAAUUCUGGCCCCGCCAGGAGAAAAGCAAUAAAAGCCUUUAAAUAUUUACACACUCAGCCAGAAAAGAGCAAACAAGCGGGCGC ..(((((((((.((((((((((......)))))).....(((((((.((....))..(((.((.......)).))).................)))))))....))))...))))))))) ( -33.30) >DroYak_CAF1 72786 120 + 1 CAGUGUCUGCUAUUGCCAUUCACGAAUAUGAAUGAAAUUUUUUGGCCCCGCCAGGAGGAAAGCAAUAAAAGCUUUUAAAUAUUUACACACUCAGCCAGAAAAGAGCAAACAAGCGGGCGC ..(((((((((.((((((((((......))))))...(((((((((.((....))..((((((.......)))))).................)))))))))..))))...))))))))) ( -37.00) >consensus CAGUGUCUGCUAUUGCCAUUCACGAAUAUGAAUGAAAAAUUUUGGCCCCGCCAGGAGAAAAGCAAUAAAAGCCUUUAAAUAUUUACACACUCAGCCAGAAAAGAGCAAACAAGCGGGCGC ..(((((((((.((((((((((......)))))).....(((((((.((....))..(((.((.......)).))).................)))))))....))))...))))))))) (-30.95 = -30.82 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:08:04 2006