
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,142,169 – 22,142,369 |
| Length | 200 |
| Max. P | 0.999543 |

| Location | 22,142,169 – 22,142,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -40.33 |
| Consensus MFE | -37.74 |
| Energy contribution | -37.22 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
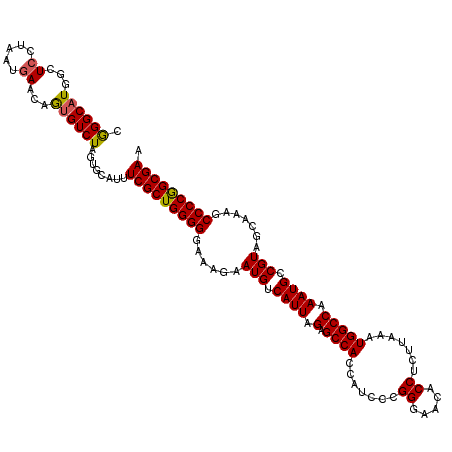
>3R_DroMel_CAF1 22142169 120 + 27905053 UGGCACUUUGUGCGCGAAUUUAUUCUUUCUUUUGCCCGCUGCAAAUUGCACCUCUGUGAUUAUGGCCACUUGGCAUCGUCUUCGCUGGGGUUUAGCUACGGCAUUUGGCCAUUUAAGAGG ..(((.((((((((((((............)))))..)).))))).))).(((((......(((((((((..((.........))..)).....((....))...)))))))...))))) ( -39.20) >DroSec_CAF1 72564 120 + 1 UGGCACUUUGUGUGCGAAUUUAUUCUUUCUUUUGCCCGCUGCAAAUUGCACCUCUGUGAUUAUGGCCACUUGGCAUCGUCUUCGCUGGGGCUUUGCUACGGCAUUUGGCCAUUUAAGAGG ..(((.((((((((((((............)))))..)).))))).))).(((((......(((((((((..((.........))..))....(((....)))..)))))))...))))) ( -38.30) >DroSim_CAF1 65571 120 + 1 UGGCACUUUGUGCGCGAAUUUAUUCUUUCUUUUGCCCGCUGCAAAUUGCACCUCUGUGAUUAUGGCCACUUGGCAUCGUCUUCGCCGGGGCUUUGCUACGGCAUUUGGCCAUUUAAGAGG ..(((.((((((((((((............)))))..)).))))).))).(((((......(((((((((((((.........))))))....(((....)))..)))))))...))))) ( -41.40) >DroEre_CAF1 67830 120 + 1 UGGCACUUUGUGCGCGAAUUUAUUCUUUCUUUUGCUCGCUGCAAAUUGCACCUCUGUGAUUAUGGCCACUUGGCAUCGUCUUCGCCGGGGCUUUGCUACGGCAUUUGGCCAUUUAGGAGG ..(((.((((((((((((............)))))..)).))))).))).(((((......(((((((((((((.........))))))....(((....)))..)))))))...))))) ( -41.40) >DroYak_CAF1 65139 120 + 1 UGGCACUUUGUGCGCGAAUUUAUUCCUUCUUUAGCUCGCUGCAAAUUGCACCUCUGUGAUUAUGGCCACUUGGCAUCGUCUUCGCCGGGGCUUUGCUACGGCAUUUGGCCAUUUAAGAGG ..(((.((((((.((((..................)))))))))).))).(((((......(((((((((((((.........))))))....(((....)))..)))))))...))))) ( -41.37) >consensus UGGCACUUUGUGCGCGAAUUUAUUCUUUCUUUUGCCCGCUGCAAAUUGCACCUCUGUGAUUAUGGCCACUUGGCAUCGUCUUCGCCGGGGCUUUGCUACGGCAUUUGGCCAUUUAAGAGG ..(((.((((((((((((............)))))..)).))))).))).(((((......(((((((...(((...)))...((((..((...))..))))...)))))))...))))) (-37.74 = -37.22 + -0.52)


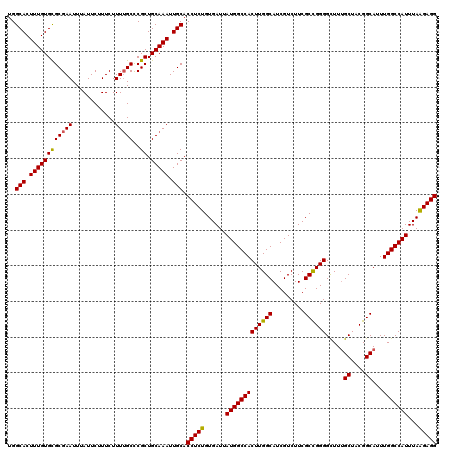
| Location | 22,142,169 – 22,142,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -28.63 |
| Energy contribution | -28.79 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22142169 120 - 27905053 CCUCUUAAAUGGCCAAAUGCCGUAGCUAAACCCCAGCGAAGACGAUGCCAAGUGGCCAUAAUCACAGAGGUGCAAUUUGCAGCGGGCAAAAGAAAGAAUAAAUUCGCGCACAAAGUGCCA (((((...(((((((...(((((.(((.......)))....)))..))....)))))))......))))).((..(((((.....))))).....(((....)))))((((...)))).. ( -30.70) >DroSec_CAF1 72564 120 - 1 CCUCUUAAAUGGCCAAAUGCCGUAGCAAAGCCCCAGCGAAGACGAUGCCAAGUGGCCAUAAUCACAGAGGUGCAAUUUGCAGCGGGCAAAAGAAAGAAUAAAUUCGCACACAAAGUGCCA ..((((..(((((.....)))))......((((.......((..(((((....)).)))..)).....(.(((.....))).)))))..))))............((((.....)))).. ( -29.70) >DroSim_CAF1 65571 120 - 1 CCUCUUAAAUGGCCAAAUGCCGUAGCAAAGCCCCGGCGAAGACGAUGCCAAGUGGCCAUAAUCACAGAGGUGCAAUUUGCAGCGGGCAAAAGAAAGAAUAAAUUCGCGCACAAAGUGCCA (((((...((((((...(((((..((...))..)))))...((........))))))))......))))).((..(((((.....))))).....(((....)))))((((...)))).. ( -32.90) >DroEre_CAF1 67830 120 - 1 CCUCCUAAAUGGCCAAAUGCCGUAGCAAAGCCCCGGCGAAGACGAUGCCAAGUGGCCAUAAUCACAGAGGUGCAAUUUGCAGCGAGCAAAAGAAAGAAUAAAUUCGCGCACAAAGUGCCA ........((((((...(((((..((...))..)))))...((........)))))))).........((..(....(((.(((((................))))))))....)..)). ( -32.09) >DroYak_CAF1 65139 120 - 1 CCUCUUAAAUGGCCAAAUGCCGUAGCAAAGCCCCGGCGAAGACGAUGCCAAGUGGCCAUAAUCACAGAGGUGCAAUUUGCAGCGAGCUAAAGAAGGAAUAAAUUCGCGCACAAAGUGCCA (((((...((((((...(((((..((...))..)))))...((........))))))))......))))).(((.((((..(((((................)))))...)))).))).. ( -32.89) >consensus CCUCUUAAAUGGCCAAAUGCCGUAGCAAAGCCCCGGCGAAGACGAUGCCAAGUGGCCAUAAUCACAGAGGUGCAAUUUGCAGCGGGCAAAAGAAAGAAUAAAUUCGCGCACAAAGUGCCA (((((...(((((((..((.(((..(...((....))...)...))).))..)))))))......))))).(((.((((..(((((................)))))...)))).))).. (-28.63 = -28.79 + 0.16)


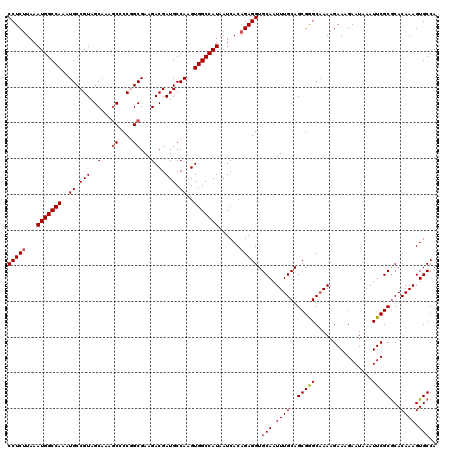
| Location | 22,142,209 – 22,142,329 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -43.10 |
| Consensus MFE | -41.24 |
| Energy contribution | -40.52 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22142209 120 + 27905053 GCAAAUUGCACCUCUGUGAUUAUGGCCACUUGGCAUCGUCUUCGCUGGGGUUUAGCUACGGCAUUUGGCCAUUUAAGAGGUGUUCCCGGGAUGGUGGCUCUAAUGACAUUCUUUCCCCCA .......((((((((......(((((((((..((.........))..)).....((....))...)))))))...))))))))....((((.((..(.((....)))...)).))))... ( -41.00) >DroSec_CAF1 72604 120 + 1 GCAAAUUGCACCUCUGUGAUUAUGGCCACUUGGCAUCGUCUUCGCUGGGGCUUUGCUACGGCAUUUGGCCAUUUAAGAGGUGUUCCCGGGAUGGUGGCUCUAAUGACAUUCUUUCCCCCA .......((((((((......(((((((((..((.........))..))....(((....)))..)))))))...))))))))....((((.((..(.((....)))...)).))))... ( -41.50) >DroSim_CAF1 65611 120 + 1 GCAAAUUGCACCUCUGUGAUUAUGGCCACUUGGCAUCGUCUUCGCCGGGGCUUUGCUACGGCAUUUGGCCAUUUAAGAGGUGUUCCCGGGAUGGUGGCUCUAAUGACAUUCUUUCCCCCA .......((((((((......(((((((((((((.........))))))....(((....)))..)))))))...))))))))....((((.((..(.((....)))...)).))))... ( -43.20) >DroEre_CAF1 67870 120 + 1 GCAAAUUGCACCUCUGUGAUUAUGGCCACUUGGCAUCGUCUUCGCCGGGGCUUUGCUACGGCAUUUGGCCAUUUAGGAGGUGCGCCCGGGAUGGUGGCUCUAAUGACAUUCUUUCCCCCA .......((((((((......(((((((((((((.........))))))....(((....)))..)))))))...))))))))....((((.((..(.((....)))...)).))))... ( -46.00) >DroYak_CAF1 65179 120 + 1 GCAAAUUGCACCUCUGUGAUUAUGGCCACUUGGCAUCGUCUUCGCCGGGGCUUUGCUACGGCAUUUGGCCAUUUAAGAGGUGUGCCCGGGAUGGUGGCUCUAAUGACAUUCUUUCCCCCA .......((((((((......(((((((((((((.........))))))....(((....)))..)))))))...))))))))....((((.((..(.((....)))...)).))))... ( -43.80) >consensus GCAAAUUGCACCUCUGUGAUUAUGGCCACUUGGCAUCGUCUUCGCCGGGGCUUUGCUACGGCAUUUGGCCAUUUAAGAGGUGUUCCCGGGAUGGUGGCUCUAAUGACAUUCUUUCCCCCA .......((((((((......(((((((...(((...)))...((((..((...))..))))...)))))))...))))))))....((((.((..(.((....)))...)).))))... (-41.24 = -40.52 + -0.72)

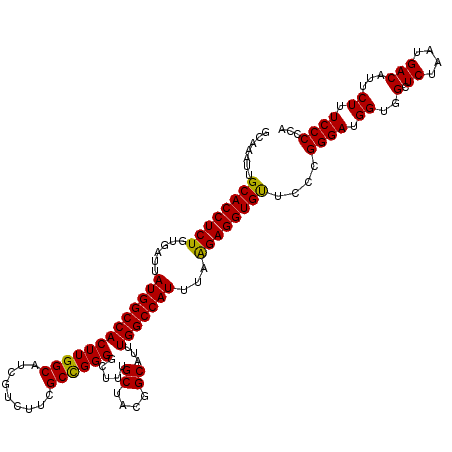
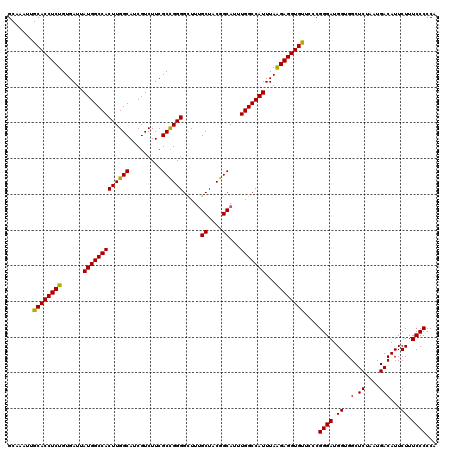
| Location | 22,142,249 – 22,142,369 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -38.32 |
| Consensus MFE | -33.10 |
| Energy contribution | -32.94 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22142249 120 - 27905053 CGGGCAUGGCUCCUAAUGAACAGUGUCUAGUGCACAUCGCUGGGGGAAAGAAUGUCAUUAGAGCCACCAUCCCGGGAACACCUCUUAAAUGGCCAAAUGCCGUAGCUAAACCCCAGCGAA .(..(((........)))..).((((.....)))).(((((((((...((.(((.((((.(.((((.......((.....)).......))))).)))).)))..))...))))))))). ( -37.24) >DroSec_CAF1 72644 120 - 1 CAGGCAUGACUCCUAAUAAACAGUGUCUAGUGCAUUUCGCUGGGGGAAAGAAUGUCAUUAGAGCCACCAUCCCGGGAACACCUCUUAAAUGGCCAAAUGCCGUAGCAAAGCCCCAGCGAA ...(((((((..............)))..))))..((((((((((........((((((((((...((.....))......))))..))))))....(((....)))...)))))))))) ( -34.44) >DroSim_CAF1 65651 120 - 1 CGGGCAUGGCUCCUAAUGAACAGUGUCUAGUGCAUUUCGCUGGGGGAAAGAAUGUCAUUAGAGCCACCAUCCCGGGAACACCUCUUAAAUGGCCAAAUGCCGUAGCAAAGCCCCGGCGAA ..((..((((((.((((((.((.(.(((((((.....))))))).)......)))))))))))))))).(((((((............(((((.....))))).((...))))))).)). ( -39.10) >DroEre_CAF1 67910 120 - 1 CGGGCUUGGCUCCUAAUGAACAAUGUCUAGUGCAUUUCGCUGGGGGAAAGAAUGUCAUUAGAGCCACCAUCCCGGGCGCACCUCCUAAAUGGCCAAAUGCCGUAGCAAAGCCCCGGCGAA ..((..((((((.((((((....(.(((((((.....))))))).)........)))))))))))))).(((((((.((.....(((...(((.....))).)))....))))))).)). ( -39.80) >DroYak_CAF1 65219 120 - 1 CGGGCAUGGCUCCUAAUGAACAGUGUCUAGUGCAUUUCGCUGGGGGAAAGAAUGUCAUUAGAGCCACCAUCCCGGGCACACCUCUUAAAUGGCCAAAUGCCGUAGCAAAGCCCCGGCGAA ((((..((((((.((((((.((.(.(((((((.....))))))).)......))))))))))))))....))))(((.((.........)))))...(((((..((...))..))))).. ( -41.00) >consensus CGGGCAUGGCUCCUAAUGAACAGUGUCUAGUGCAUUUCGCUGGGGGAAAGAAUGUCAUUAGAGCCACCAUCCCGGGAACACCUCUUAAAUGGCCAAAUGCCGUAGCAAAGCCCCGGCGAA .((((((...((.....))...))))))........(((((((((......(((.((((.(.((((.......((.....)).......))))).)))).))).......))))))))). (-33.10 = -32.94 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:42 2006