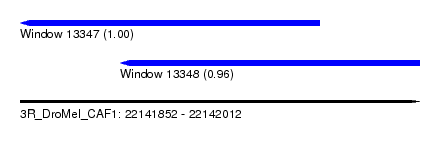
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,141,852 – 22,142,012 |
| Length | 160 |
| Max. P | 0.997566 |

| Location | 22,141,852 – 22,141,972 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -32.58 |
| Energy contribution | -33.34 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22141852 120 - 27905053 GAGAUUGUAAUUAGUUUUUGUUGCUCCUGGGUUUAACUGGAGGGAAACUUUCAAAUUGAACCAAAGACGGCGGCAAAGGGAUGGAAUAUUUUGCGGUUCCAACUUUUACUCACUGGUCUU ((((((((((..(((((((((((((....(((((((.((((((....))))))..)))))))......)))))))))(((((.(((....)).).))))))))).)))).....)))))) ( -34.70) >DroSec_CAF1 72247 120 - 1 GAGAUUGUAAUUAGUUUUUGUUGCUCCUGGGUUUAACUGGAGGGAAACUUUCAAAUUGAACCAAAGACGGCGGCAAAGGGAUGGAAUAUUUUGCGGUUCCAACUUUUACACACUGCUCUU (((..(((((..(((((((((((((....(((((((.((((((....))))))..)))))))......)))))))))(((((.(((....)).).))))))))).))))).....))).. ( -36.50) >DroSim_CAF1 65254 120 - 1 GAGAUUGUAAUUAGUUUUUGUUGCUCCUGGGUUUAACUGGAGGGAAACUUUCAAAUUGAACCAAAGACGGCGGCAAAGGGAUGGAAUAUUUUGCGGUUCCAACUUUUACACACUGGUCUU ((((((((((..(((((((((((((....(((((((.((((((....))))))..)))))))......)))))))))(((((.(((....)).).))))))))).))))).....))))) ( -35.80) >DroEre_CAF1 67515 120 - 1 GAGAUUGUAAUUAGUUUUUGUUGCUUCUGGGUUUAACUGGAGGGAAACUUUGAAAUUGAACCAAAGACGCCGGCAAAGGGAUGGAAUAUUUUGCGGUCCCAACUUUUACACAUUGGUCUU ((((((((((..((((((((((((.(((.(((((((.(.((((....)))).)..)))))))..))).).)))))))(((((.(((....)).).))))))))).))))).....))))) ( -33.60) >DroYak_CAF1 64827 120 - 1 GAGAUUGUAAUUAGUUUUUGUUGCUUCUGGGUUUAACUGGAGGGAAACUUUCAAAUUGAACCAAAGACGCCGGCAAAGGGAUGGAAUAUUUUGCGGUCCCAACUUUUACACACUGGUCUU ((((((((((..((((((((((((.(((.(((((((.((((((....))))))..)))))))..))).).)))))))(((((.(((....)).).))))))))).))))).....))))) ( -36.80) >consensus GAGAUUGUAAUUAGUUUUUGUUGCUCCUGGGUUUAACUGGAGGGAAACUUUCAAAUUGAACCAAAGACGGCGGCAAAGGGAUGGAAUAUUUUGCGGUUCCAACUUUUACACACUGGUCUU ((((((((((..(((((((((((((....(((((((.((((((....))))))..)))))))......)))))))))(((((.(((....)).).))))))))).))))).....))))) (-32.58 = -33.34 + 0.76)

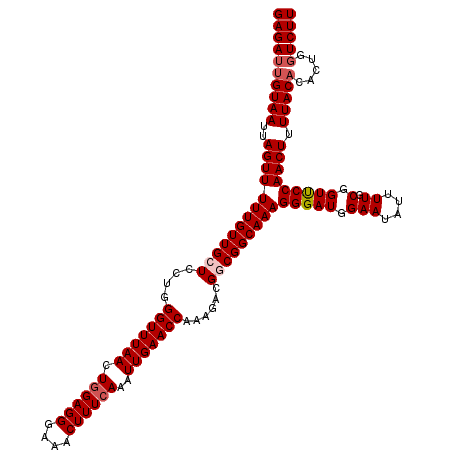
| Location | 22,141,892 – 22,142,012 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.16 |
| Mean single sequence MFE | -32.33 |
| Consensus MFE | -27.90 |
| Energy contribution | -28.50 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22141892 120 - 27905053 CAUUUAGCUUUAAGUUGGCCGAGUGUUGAAAAAACAACUUGAGAUUGUAAUUAGUUUUUGUUGCUCCUGGGUUUAACUGGAGGGAAACUUUCAAAUUGAACCAAAGACGGCGGCAAAGGG ......(((.......)))((((((((.....))).))))).............(((((((((((....(((((((.((((((....))))))..)))))))......))))))))))). ( -32.50) >DroSec_CAF1 72287 120 - 1 CAUUUAGAUUUAAGUUGGCCGAGUGUUGAAAAAACAACUUGAGAUUGUAAUUAGUUUUUGUUGCUCCUGGGUUUAACUGGAGGGAAACUUUCAAAUUGAACCAAAGACGGCGGCAAAGGG ........(((((((((.................)))))))))...........(((((((((((....(((((((.((((((....))))))..)))))))......))))))))))). ( -32.53) >DroSim_CAF1 65294 120 - 1 CAUUUAGCUUUAAGUUGGCCGAGUGUUGAAAAAACAACUUGAGAUUGUAAUUAGUUUUUGUUGCUCCUGGGUUUAACUGGAGGGAAACUUUCAAAUUGAACCAAAGACGGCGGCAAAGGG ......(((.......)))((((((((.....))).))))).............(((((((((((....(((((((.((((((....))))))..)))))))......))))))))))). ( -32.50) >DroEre_CAF1 67555 117 - 1 CAUUUAGCUUUAAG---GCCGAGUGCUGAAAAAACCACUUGAGAUUGUAAUUAGUUUUUGUUGCUUCUGGGUUUAACUGGAGGGAAACUUUGAAAUUGAACCAAAGACGCCGGCAAAGGG .......((((...---((((.(((((..............(((..(((((........))))).))).(((((((.(.((((....)))).)..)))))))..)).))))))).)))). ( -28.70) >DroYak_CAF1 64867 117 - 1 CAUUUAGCUUUAAG---GCCGAGUGUUGAAAAAACAACUUGAGAUUGUAAUUAGUUUUUGUUGCUUCUGGGUUUAACUGGAGGGAAACUUUCAAAUUGAACCAAAGACGCCGGCAAAGGG .......((((...---((((.(((((.......((((..(((((((....))))))).))))......(((((((.((((((....))))))..)))))))...))))))))).)))). ( -35.40) >consensus CAUUUAGCUUUAAGUUGGCCGAGUGUUGAAAAAACAACUUGAGAUUGUAAUUAGUUUUUGUUGCUCCUGGGUUUAACUGGAGGGAAACUUUCAAAUUGAACCAAAGACGGCGGCAAAGGG .................((((.((.((.......((((..(((((((....))))))).))))......(((((((.((((((....))))))..))))))).)).))..))))...... (-27.90 = -28.50 + 0.60)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:37 2006