
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,133,159 – 22,133,249 |
| Length | 90 |
| Max. P | 0.947903 |

| Location | 22,133,159 – 22,133,249 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.39 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -17.08 |
| Energy contribution | -16.83 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.947903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
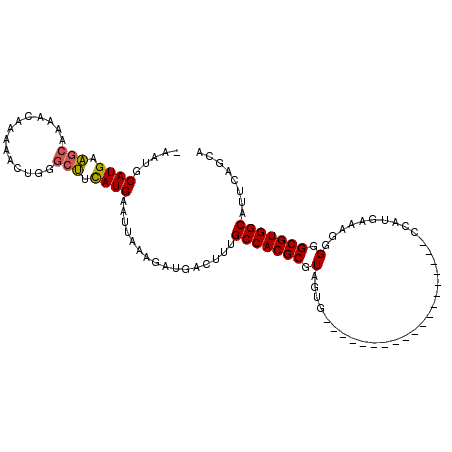
>3R_DroMel_CAF1 22133159 90 + 27905053 -AAUGCAUGAAGCAAAACAAAAACUGGGCUUCAUGAAUUAAAGAUGACUUUGCCACGCGUAGUG-------------------CCAUGAAAGGGGGCGUGGCAUUCGGCA -....((((((((....((.....)).)))))))).......(.(((...((((((((.(....-------------------((......))).)))))))).))).). ( -27.70) >DroPse_CAF1 89392 104 + 1 GAAUGCAUGCGGCAA-----AAACUU-GCCUCAUGAAUUAAAGAUGACUUUGCCACGCGUUUGGCUGCUCCGCAGCUGCCAAGCCGUGACAGUGGGCGUGGCCGGCAACA .(((.((((.(((((-----....))-))).)))).)))............((((((((((((((.(((....))).)))))))((......)).))))))).(....). ( -38.90) >DroSec_CAF1 63536 90 + 1 -AAUGCAUGAAGCAAAACAAAAACUGGGCUUCAUGAAUUAAAGAUGACUUUGCCACGCGUAGUG-------------------CCAUGAAAGGGGGCGUGGCAUUCGGCA -....((((((((....((.....)).)))))))).......(.(((...((((((((.(....-------------------((......))).)))))))).))).). ( -27.70) >DroSim_CAF1 56491 90 + 1 -AAUGCAUGAAGCAAAACAAAAACUGGGCUUCAUGAAUUAAAGAUGACUUUGCCACGCGUAGUG-------------------CCAUGAAAGGGGGCGUGGCAUUCGGCA -....((((((((....((.....)).)))))))).......(.(((...((((((((.(....-------------------((......))).)))))))).))).). ( -27.70) >DroAna_CAF1 103882 98 + 1 -AAUGCAUGCACGAGACAAAAAACUUGGCUUUAUGAAUUAAAGAUGACUUCGCCACGCGUUCUG-----------CUGCCAAGCCCUGACAGGGGGCGUGGCAUCCAGCA -..(((..((.((((........))))))......................(((((((.(((((-----------(.((...))...).))))).))))))).....))) ( -28.30) >DroPer_CAF1 90066 104 + 1 GAAUGCAUGCGGCAA-----AAACUU-GCCUCAUGAAUUAAAGAUGACUUUGCCACGCGUUUGGCUGCUCCGCAGCUGCCAAGCCGUGACAGUGGGCGUGGCCGGCAACA .(((.((((.(((((-----....))-))).)))).)))............((((((((((((((.(((....))).)))))))((......)).))))))).(....). ( -38.90) >consensus _AAUGCAUGAAGCAAAACAAAAACUGGGCUUCAUGAAUUAAAGAUGACUUUGCCACGCGUAGUG___________________CCAUGAAAGGGGGCGUGGCAUUCAGCA .....((((.(((..............))).))))................(((((((.(.................................).)))))))........ (-17.08 = -16.83 + -0.25)

| Location | 22,133,159 – 22,133,249 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.39 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -15.45 |
| Energy contribution | -15.37 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
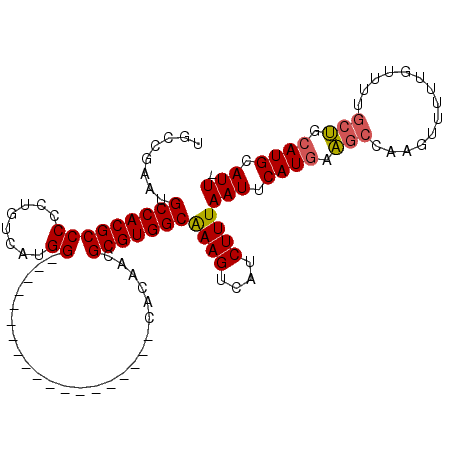
>3R_DroMel_CAF1 22133159 90 - 27905053 UGCCGAAUGCCACGCCCCCUUUCAUGG-------------------CACUACGCGUGGCAAAGUCAUCUUUAAUUCAUGAAGCCCAGUUUUUGUUUUGCUUCAUGCAUU- (((.(((((((((((..((......))-------------------......)))))))((((....)))).))))(((((((..............))))))))))..- ( -24.94) >DroPse_CAF1 89392 104 - 1 UGUUGCCGGCCACGCCCACUGUCACGGCUUGGCAGCUGCGGAGCAGCCAAACGCGUGGCAAAGUCAUCUUUAAUUCAUGAGGC-AAGUUU-----UUGCCGCAUGCAUUC (((..(((((((.(((.........))).)))).(((((...)))))....)).)..)))...........(((.((((.(((-((....-----))))).)))).))). ( -34.80) >DroSec_CAF1 63536 90 - 1 UGCCGAAUGCCACGCCCCCUUUCAUGG-------------------CACUACGCGUGGCAAAGUCAUCUUUAAUUCAUGAAGCCCAGUUUUUGUUUUGCUUCAUGCAUU- (((.(((((((((((..((......))-------------------......)))))))((((....)))).))))(((((((..............))))))))))..- ( -24.94) >DroSim_CAF1 56491 90 - 1 UGCCGAAUGCCACGCCCCCUUUCAUGG-------------------CACUACGCGUGGCAAAGUCAUCUUUAAUUCAUGAAGCCCAGUUUUUGUUUUGCUUCAUGCAUU- (((.(((((((((((..((......))-------------------......)))))))((((....)))).))))(((((((..............))))))))))..- ( -24.94) >DroAna_CAF1 103882 98 - 1 UGCUGGAUGCCACGCCCCCUGUCAGGGCUUGGCAG-----------CAGAACGCGUGGCGAAGUCAUCUUUAAUUCAUAAAGCCAAGUUUUUUGUCUCGUGCAUGCAUU- .......(((((.((((.......)))).)))))(-----------((...((((.(((((((....(((((.....))))).......))))))).))))..)))...- ( -31.60) >DroPer_CAF1 90066 104 - 1 UGUUGCCGGCCACGCCCACUGUCACGGCUUGGCAGCUGCGGAGCAGCCAAACGCGUGGCAAAGUCAUCUUUAAUUCAUGAGGC-AAGUUU-----UUGCCGCAUGCAUUC (((..(((((((.(((.........))).)))).(((((...)))))....)).)..)))...........(((.((((.(((-((....-----))))).)))).))). ( -34.80) >consensus UGCCGAAUGCCACGCCCCCUGUCAUGG___________________CACAACGCGUGGCAAAGUCAUCUUUAAUUCAUGAAGCCAAGUUUUUGUUUUGCUGCAUGCAUU_ ........(((((((((........)).........................)))))))((((....))))(((.((((.(((..............))).)))).))). (-15.45 = -15.37 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:23 2006