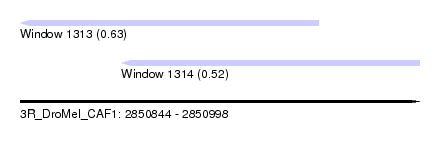
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,850,844 – 2,850,998 |
| Length | 154 |
| Max. P | 0.634515 |

| Location | 2,850,844 – 2,850,959 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.89 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -14.28 |
| Energy contribution | -14.33 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
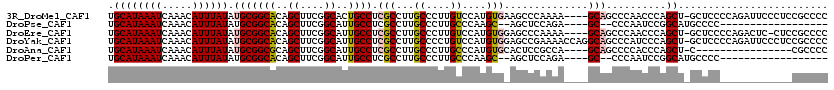
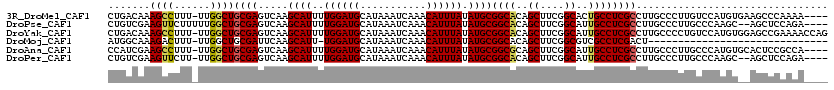
>3R_DroMel_CAF1 2850844 115 - 27905053 UGCAUAAAUCAAACAUUUAUAUGCGGCACAGCUUCGGCACUGCCUCGCCUUGCCCUUGUCCAUGUGAAGCCCAAAA----GCAGCCCAACCCAGCU-GCUCCCCAGAUUCCCUCCGCCCC .(((((.............)))))(((...(((((((((..((...))..))))(........).))))).....(----(((((........)))-)))...............))).. ( -26.72) >DroPse_CAF1 191288 94 - 1 UGCAUAAAUCAAACAUUUAUAUGCGGCACAGCUUCGGCAUUGCCUCGCCUUGCCCUUGCCCAAGC--AGCUCCAGA----GC--CCCAAUCCGGCAUGCCCC------------------ .(((((.............)))))((((..((....))..))))..((..((((..(((....))--)(((....)----))--........)))).))...------------------ ( -24.92) >DroEre_CAF1 128862 114 - 1 UGCAUAAAUCAAACAUUUAUAUGCGGCACAGCUUCGGCAUUGCCUCGCCUUGCCCUUGUCCAUGUGGAGCCCAAAA----GCAGCCCAACCCAGCU-GCUCCCCAGACUC-CUCCGCCCC .(((((.............)))))((((((((...((((..((...))..))))...))...)))(((((.....(----(((((........)))-))).....).)))-)...))).. ( -29.52) >DroYak_CAF1 138943 119 - 1 UGCAUAAAUCAAACAUUUAUAUGCGGCACAGCUUCGGCAUUGCCUCGCCUUGCCCCUGUCCAUGUGGAGCCGAAAACCAGGCAGCCCAUCCCAGCU-GCUCCCCAGAUUCCCUCCGCCCC .(((((.............)))))((.((((....((((..((...))..)))).))))))..((((((..(((.....((((((........)))-))).......))).))))))... ( -32.32) >DroAna_CAF1 118282 99 - 1 UGCAUAAAUCAAACAUUUAUAUGCGGCGCAGCUUCGGCAUUGCCUCGCCUUGCCCUUGCCCAUGUGCACUCCGCCA----GCAGCCCCACCCAGCU-C----------------CGCCCC .(((((.............)))))((((.((((..((((..((...))..))))...((...((.((.....))))----...)).......))))-.----------------)))).. ( -25.22) >DroPer_CAF1 187305 94 - 1 UGCAUAAAUCAAACAUUUAUAUGCGGCACAGCUUCGGCAUUGCCUCGCCUUGCCCUUGCCCAAGC--AGCUCCAGA----GC--CCCAAUCCGGCAUGCCCC------------------ .(((((.............)))))((((..((....))..))))..((..((((..(((....))--)(((....)----))--........)))).))...------------------ ( -24.92) >consensus UGCAUAAAUCAAACAUUUAUAUGCGGCACAGCUUCGGCAUUGCCUCGCCUUGCCCUUGCCCAUGUG_AGCCCAAAA____GCAGCCCAACCCAGCU_GCUCC____________CGCCCC .((((((((.....)))))).(((((((..((....))..)))).(((...((....))....)))..............)))..........))......................... (-14.28 = -14.33 + 0.06)
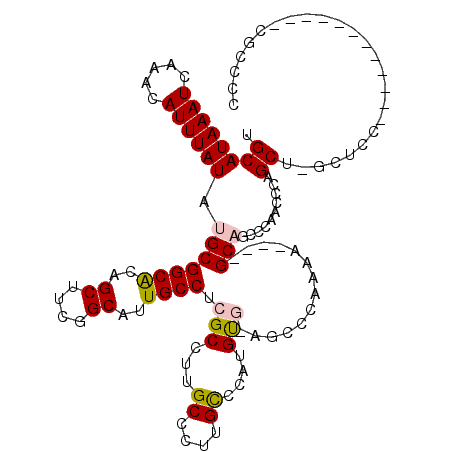
| Location | 2,850,883 – 2,850,998 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.48 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -22.65 |
| Energy contribution | -22.65 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
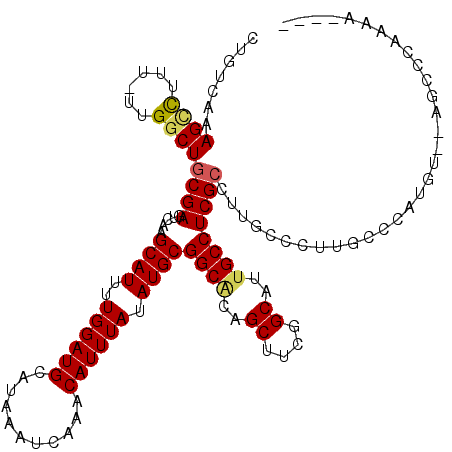
>3R_DroMel_CAF1 2850883 115 - 27905053 CUGACAAAGCCUUU-UUGGCUGCGAGUCAAGCAUUUUGGAUGCAUAAAUCAAACAUUUAUAUGCGGCACAGCUUCGGCACUGCCUCGCCUUGCCCUUGUCCAUGUGAAGCCCAAAA---- ..(((((((((...-..))))(((((.(..((((..((((((...........)))))).))))((((..((....))..))))..).)))))..)))))................---- ( -32.80) >DroPse_CAF1 191308 114 - 1 CUGUCGAAGUUCUUUUUGGCUGCGAGUCAAGCAUUUUGGAUGCAUAAAUCAAACAUUUAUAUGCGGCACAGCUUCGGCAUUGCCUCGCCUUGCCCUUGCCCAAGC--AGCUCCAGA---- (((....(((((((...(((.(((((.(..((((..((((((...........)))))).))))((((..((....))..))))..).)))))....))).))).--)))).))).---- ( -30.90) >DroYak_CAF1 138982 119 - 1 CUGACAAAGCCUUU-UUGGCUGCGAGUCAAGCAUUUUGGAUGCAUAAAUCAAACAUUUAUAUGCGGCACAGCUUCGGCAUUGCCUCGCCUUGCCCCUGUCCAUGUGGAGCCGAAAACCAG ...........(((-(((((.(((((.(..((((..((((((...........)))))).))))((((..((....))..))))..).))))).....(((....))))))))))).... ( -34.30) >DroMoj_CAF1 196031 88 - 1 AUGGCAAAGACUUU-UUGGCUGCGAUUCAAGCAUU-UGGAUGCAUAAAUCAAACAUUUAUAUGCGGCACAGCUUCGGCGUCGCCUCGACU------------------------------ ..(((...(((...-.((((((((......((((.-...))))((((((.....)))))).)))))).))((....))))))))......------------------------------ ( -24.20) >DroAna_CAF1 118305 115 - 1 CCAUCGAAGCCUUU-UUGGCUGCGAGUCAAGCAUUUUGGAUGCAUAAAUCAAACAUUUAUAUGCGGCGCAGCUUCGGCAUUGCCUCGCCUUGCCCUUGCCCAUGUGCACUCCGCCA---- ...(((.((((...-..)))).)))............(((((((((.......(((....))).((.((((....((((..((...))..)))).)))))).)))))).)))....---- ( -31.90) >DroPer_CAF1 187325 113 - 1 CUGUCGAAGUUCUU-UUGGCUGCGAGUCAAGCAUUUUGGAUGCAUAAAUCAAACAUUUAUAUGCGGCACAGCUUCGGCAUUGCCUCGCCUUGCCCUUGCCCAAGC--AGCUCCAGA---- (((....((((.((-(.(((.(((((.(..((((..((((((...........)))))).))))((((..((....))..))))..).)))))....))).))).--)))).))).---- ( -31.40) >consensus CUGUCAAAGCCUUU_UUGGCUGCGAGUCAAGCAUUUUGGAUGCAUAAAUCAAACAUUUAUAUGCGGCACAGCUUCGGCAUUGCCUCGCCUUGCCCUUGCCCAUGU__AGCCCAAAA____ .......((((......))))((((.....((((..((((((...........)))))).))))((((..((....))..))))))))................................ (-22.65 = -22.65 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:05 2006