
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,103,498 – 22,103,589 |
| Length | 91 |
| Max. P | 0.620072 |

| Location | 22,103,498 – 22,103,589 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -19.13 |
| Consensus MFE | -15.72 |
| Energy contribution | -15.75 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22103498 91 + 27905053 CCGCAGAGGCAACAACUUUUGUUAAACAUUUGUUACCAGCAGCC--------AUACCACUGGCAGCCCCAACAACCAUCACUUACCAGUGGAAACUAAU ((((...((.(((((....((.....)).))))).)).((.(((--------(......)))).)).....................))))........ ( -17.20) >DroSec_CAF1 26345 90 + 1 CCGCAGAGGCAACAACUUUUGUUAAACAUUUGUUACCAGCAGCC--------AUACCACUGGCAGCC-CAACAACCAUCACUUACCAGUGGAAACUAAU ((((...((.(((((....((.....)).))))).)).((.(((--------(......)))).)).-...................))))........ ( -17.20) >DroSim_CAF1 26187 91 + 1 CCGCAGAGGCAACAACUUUUGUUAAACAUUUGUUACCAGCAGCC--------AUACCACUGGCAGCCCCAACAACCAUCACUUACCAGUGGAAACUAAU ((((...((.(((((....((.....)).))))).)).((.(((--------(......)))).)).....................))))........ ( -17.20) >DroEre_CAF1 26858 99 + 1 CCGCAGAGGCAACAACUUUUGUUAAACAUUUGUUACCAGCAGCCAUACUGCCAUACCACUGGCAGCCCCAACAGCCAUCACUUACCAGUGGAAACUAAU ..((((.(((((((.....)))).......(((.....))))))...))))....(((((((......................)))))))........ ( -20.95) >DroYak_CAF1 25868 99 + 1 CCGCAGAGGCAACAACUUUUGUUAAACAUUUGUUACCAGCAGCCAUGCUGCCAUACCACUGGCAGCCCCAACAGCCAUCACUUACCAGUGGAAACUAAU ((((...(((((((.....)))).......(((.....))))))..(((((((......))))))).....................))))........ ( -24.10) >DroAna_CAF1 27059 91 + 1 UCGCAGUGGCAGGAACUUUUGCUAAACAUUUGUUACCAGCAGCA--------AUACCACUGGCACUACCAACAGCCAUCACUUACCAGUGGAAACUAAU ..((..(((((((....)))))))......(((.....))))).--------...(((((((......................)))))))........ ( -18.15) >consensus CCGCAGAGGCAACAACUUUUGUUAAACAUUUGUUACCAGCAGCC________AUACCACUGGCAGCCCCAACAACCAUCACUUACCAGUGGAAACUAAU ..(((((((......))))))).......((((.....)))).............(((((((......................)))))))........ (-15.72 = -15.75 + 0.03)

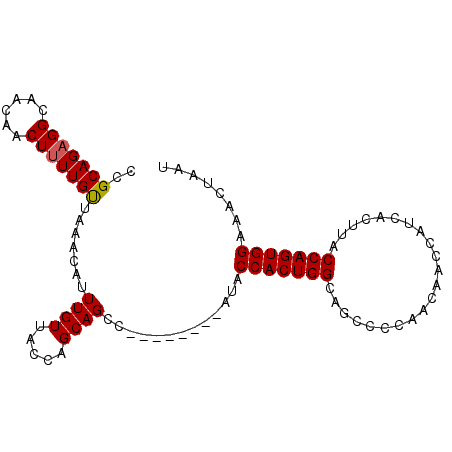
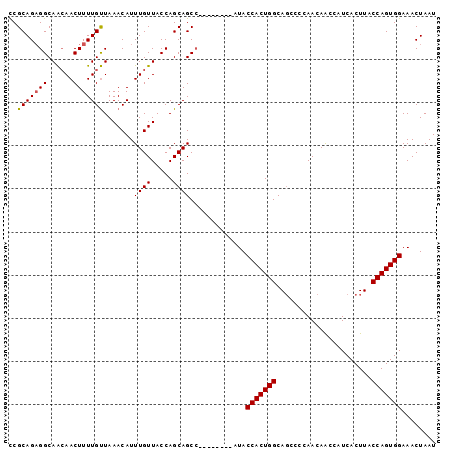
| Location | 22,103,498 – 22,103,589 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -24.50 |
| Energy contribution | -25.03 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22103498 91 - 27905053 AUUAGUUUCCACUGGUAAGUGAUGGUUGUUGGGGCUGCCAGUGGUAU--------GGCUGCUGGUAACAAAUGUUUAACAAAAGUUGUUGCCUCUGCGG ...((((.(((((((((.((..(.......)..)))))))))))...--------))))((.((((((((.((.....))....))))))))...)).. ( -28.30) >DroSec_CAF1 26345 90 - 1 AUUAGUUUCCACUGGUAAGUGAUGGUUGUUG-GGCUGCCAGUGGUAU--------GGCUGCUGGUAACAAAUGUUUAACAAAAGUUGUUGCCUCUGCGG ...((((.(((((((((.(((((....))).-.)))))))))))...--------))))((.((((((((.((.....))....))))))))...)).. ( -28.40) >DroSim_CAF1 26187 91 - 1 AUUAGUUUCCACUGGUAAGUGAUGGUUGUUGGGGCUGCCAGUGGUAU--------GGCUGCUGGUAACAAAUGUUUAACAAAAGUUGUUGCCUCUGCGG ...((((.(((((((((.((..(.......)..)))))))))))...--------))))((.((((((((.((.....))....))))))))...)).. ( -28.30) >DroEre_CAF1 26858 99 - 1 AUUAGUUUCCACUGGUAAGUGAUGGCUGUUGGGGCUGCCAGUGGUAUGGCAGUAUGGCUGCUGGUAACAAAUGUUUAACAAAAGUUGUUGCCUCUGCGG ..(((((..((((....))))..))))).....(((((((......)))))))....((((.((((((((.((.....))....))))))))...)))) ( -32.90) >DroYak_CAF1 25868 99 - 1 AUUAGUUUCCACUGGUAAGUGAUGGCUGUUGGGGCUGCCAGUGGUAUGGCAGCAUGGCUGCUGGUAACAAAUGUUUAACAAAAGUUGUUGCCUCUGCGG ..(((((..((((....))))..))))).....(((((((......)))))))....((((.((((((((.((.....))....))))))))...)))) ( -34.90) >DroAna_CAF1 27059 91 - 1 AUUAGUUUCCACUGGUAAGUGAUGGCUGUUGGUAGUGCCAGUGGUAU--------UGCUGCUGGUAACAAAUGUUUAGCAAAAGUUCCUGCCACUGCGA ..(((((..((((....))))..))))).((((((((((((..(...--------..)..)))))).....((.....)).......))))))...... ( -26.90) >consensus AUUAGUUUCCACUGGUAAGUGAUGGCUGUUGGGGCUGCCAGUGGUAU________GGCUGCUGGUAACAAAUGUUUAACAAAAGUUGUUGCCUCUGCGG ........(((((((((.(((((....)))...))))))))))).............((((.((((((((.((.....))....))))))))...)))) (-24.50 = -25.03 + 0.53)

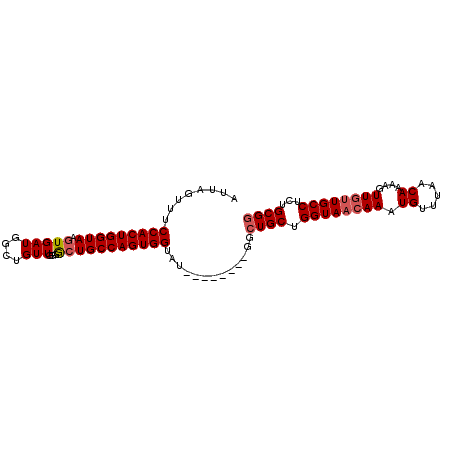
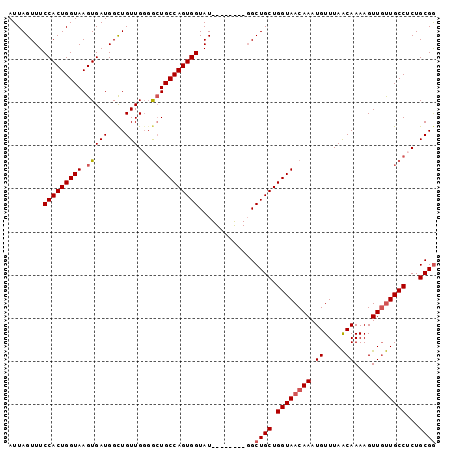
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:06 2006