
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,095,853 – 22,095,950 |
| Length | 97 |
| Max. P | 0.864150 |

| Location | 22,095,853 – 22,095,950 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 79.43 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -19.62 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22095853 97 + 27905053 ---CUGGCAUUUUUCUAUUGUUCACCGGGCUGUCCCCUAAAGGAUGCUCAGAUACUCUGACAAAGAGCGUUAGACACU--UUCGAA--AAAGG------GGAGAGCAAUU ---((((.................))))(((.((((((...(((((.((.((..((((.....))))..)).))))..--)))...--..)))------))).))).... ( -26.23) >DroSec_CAF1 18663 97 + 1 ---CUGGCAUUUUUCUAUUGUUCACCGGGCUGUCCCCUAAAGGAUGCUCAGAUACUCUGACAAAGAGUGUUACGCAAU--UUCGAG--AAAGG------GGAGAGCAAUU ---((((.................))))(((.((((((...((((((...((((((((.....))))))))..)))..--)))...--..)))------))).))).... ( -31.03) >DroSim_CAF1 18634 97 + 1 ---CUGGCAUUUUUCUAUUGUUCACCGGGCUGUCCCCUAAAGGAUGCUCAGAUACUCUGACAAAGAGUGUUAGACACU--UUCGAA--AAAGG------GGAGAGCAAUU ---((((.................))))(((.((((((...(((((.((.((((((((.....)))))))).))))..--)))...--..)))------))).))).... ( -30.33) >DroEre_CAF1 18933 98 + 1 ---CUGGCAUUUUUCUAUUGUUCACCGGGCUGUCCCCCA-AGGAUGCUUAGAUACUCUGACAAAGAGUGUUAGAAACU--UUCAGAAUGAAGG------GGUGAGCAAUU ---.............((((((((((((((.((((....-.)))))))).((((((((.....)))))))).....((--(((.....)))))------)))))))))). ( -33.10) >DroYak_CAF1 17930 99 + 1 ---CUGGCAUUUUUCUAUUGUUCAUCGGGCUGUCCCCUAAAGGAUGCUUAGAUACUCUGACAAAGGGUGUUGGAAACU--UUCAAAUUGAAGG------GGUGAGCAAUU ---.............((((((((((((((.((((......)))))))).((((((((.....)))))))).....((--(((.....)))))------)))))))))). ( -29.80) >DroPer_CAF1 51768 109 + 1 UCUGUGGUAUUUUUCAAUUGUUCAUCGUUUUGGCCUCUGAAGAAUGUGUUGAUACUCUGCCGCAGAGACUCAAAAAAUGUUUCCAAG-AAAGUUCUUUGACUGACUAAAG (((((((((....((((((((((.(((..........))).))))).))))).....)))))))))...(((((.(((.((((...)-)))))).))))).......... ( -20.70) >consensus ___CUGGCAUUUUUCUAUUGUUCACCGGGCUGUCCCCUAAAGGAUGCUCAGAUACUCUGACAAAGAGUGUUAGAAACU__UUCGAA__AAAGG______GGAGAGCAAUU ................(((((((.((((((.((((......)))))))).((((((((.....))))))))............................)).))))))). (-19.62 = -20.10 + 0.48)

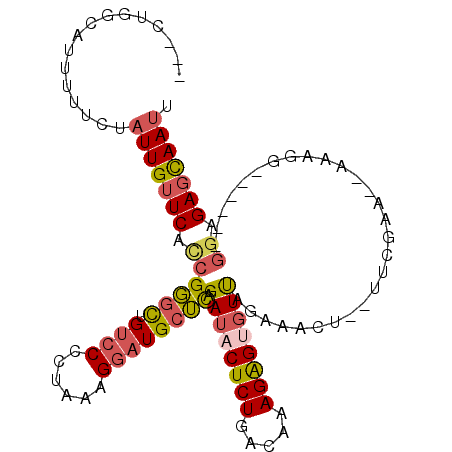

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:02 2006