
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,093,545 – 22,093,714 |
| Length | 169 |
| Max. P | 0.999538 |

| Location | 22,093,545 – 22,093,657 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.71 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -20.92 |
| Energy contribution | -20.54 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.759845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
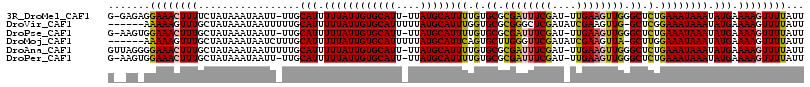
>3R_DroMel_CAF1 22093545 112 + 27905053 G-GAGAGGAAACUUUUCUAUAAAUAAUU-UUGCAUUUUUAUUGUGCAUU-UUAUGCAUUUUGUGCGCGAUUUCGAU-UUGAAGUUGGGCUCUGAAAUAAAUAUGAAAAGUUUUAUU (-(((((....)))))).......((((-((.(((.((((((((((((.-..))))))...(.((.(((((((...-..))))))).)).)...)))))).))).))))))..... ( -27.70) >DroVir_CAF1 81885 109 + 1 ------AAAAAGUUUGCUAUAAAUAAUUUUUGCAUUUUUAUUGUGCAUUUUUAUGCAUUUGGUGCGCGGGCUCGAUAUCGAAGUUG-GCUCGGAAAUAAAUAUGAAAAGUUUUAUU ------.((((.(((..((((..........(((((......((((((....))))))..))))).(((((.((((......))))-)))))........)))).))).))))... ( -22.40) >DroPse_CAF1 47961 112 + 1 G-AAGUGGAAACUUUGCUAUAAAUAAUU-UUGCAUUUUUAUUGUGCAUU-UUAUGCAUUUUGUGCGCGAUUUCGAU-UUGAAGUUGGGCUCUGAAAUAAAUAUGAAAAGUUUUAUU (-((((....))))).........((((-((.(((.((((((((((((.-..))))))...(.((.(((((((...-..))))))).)).)...)))))).))).))))))..... ( -23.90) >DroMoj_CAF1 83161 109 + 1 ------AAAAAGUUUGCUAUAAAUAAUCUUUGCAUUUUUAUUGUGCAUUUUUAUGCAUUCAGUGCUUGGGUUCGAUAUCGAAGUUA-GCUUGGAAAUAAAUAUGAAAAGUUUUAUU ------...............(((((.((((.(((.((((((.(((((....)))))(((((.(((.((.((((....)))).)))-))))))))))))).))).))))..))))) ( -21.90) >DroAna_CAF1 11898 114 + 1 GUUAGGGGAAACUUUGCUAUAAAUAAUUUUUGCAUUUUUAUUGUGCAUU-UUAUGCAUUUUGUGCGCGAUUUCGAU-UUGAAGUUGGGCUCUGAAAUAAAUAUGAAAAGUUUUAUU ..(((((....)))))........(((((((.(((.((((((((((((.-..))))))...(.((.(((((((...-..))))))).)).)...)))))).))))))))))..... ( -24.70) >DroPer_CAF1 49618 112 + 1 G-AAGUGGAAACUUUGCUAUAAAUAAUU-UUGCAUUUUUAUUGUGCAUU-UUAUGCAUUUUGUGCGCGAUUUCGAU-UUGAAGUUGGGCUCUGAAAUAAAUAUGAAAAGUUUUAUU (-((((....))))).........((((-((.(((.((((((((((((.-..))))))...(.((.(((((((...-..))))))).)).)...)))))).))).))))))..... ( -23.90) >consensus G__AG_GGAAACUUUGCUAUAAAUAAUU_UUGCAUUUUUAUUGUGCAUU_UUAUGCAUUUUGUGCGCGAUUUCGAU_UUGAAGUUGGGCUCUGAAAUAAAUAUGAAAAGUUUUAUU .......((((((((.................(((.((((((((((((....))))))((.(.((.((((((((....)))))))).)).).)))))))).))).))))))))... (-20.92 = -20.54 + -0.39)


| Location | 22,093,580 – 22,093,688 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.28 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -18.74 |
| Energy contribution | -17.88 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22093580 108 + 27905053 UUAUUGUGCAUU-UUAUGCAUUUUGUGCGCGAUUUCGAU-UUGAAGUUGGGCUCUGAAAUAAAUAUGAAAAGUU-UUAUUUAUGUUUG--UGGGG------UGGCAAAGCCCGGAAAAA .((..((((((.-..))))))..)).((.(((((((...-..))))))).))(((...(((((((((((.....-...))))))))))--).(((------(......))))))).... ( -27.30) >DroVir_CAF1 81916 110 + 1 UUAUUGUGCAUUUUUAUGCAUUUGGUGCGCGGGCUCGAUAUCGAAGUUG-GCUCGGAAAUAAAUAUGAAAAGUU-UUAUUUAUGUUUG--CGGCCA-----GGACUAAGUCGUGGCUAG .....((((((....)))))).....((.(((((.((((......))))-))))).....(((((((((.....-...))))))))))--)(((((-----.(((...))).))))).. ( -31.80) >DroPse_CAF1 47996 108 + 1 UUAUUGUGCAUU-UUAUGCAUUUUGUGCGCGAUUUCGAU-UUGAAGUUGGGCUCUGAAAUAAAUAUGAAAAGUU-UUAUUUAUGUUUGCGUGGGG------UGGCUGUGGCUGUGGC-- ........((((-((((((((((((.((.(((((((...-..))))))).)).).)))))(((((((((.....-...)))))))))))))))))------))((..(....)..))-- ( -27.20) >DroWil_CAF1 93159 111 + 1 UUAUUGUGCAUU-UUAUGCAUUUUGUGUACGAUUUCGAU-UUGAAGUUG-GCUCUGAAAUAAAUAUGAAAAGUUUUUAUUCAUGUUUG--CCGUUUAGA--GGGAAGUACCAGAGGA-G .....((((((.-..))))))((((.((((((((((...-..)))))).-.(((((((.((((((((((.........))))))))))--...))))))--)....))))))))...-. ( -27.20) >DroMoj_CAF1 83192 110 + 1 UUAUUGUGCAUUUUUAUGCAUUCAGUGCUUGGGUUCGAUAUCGAAGUUA-GCUUGGAAAUAAAUAUGAAAAGUU-UUAUUUAUGUUUG--CCACCC-----GGACUAAGCCGUGGCUAG ......(((((....)))))(((((.(((.((.((((....)))).)))-)))))))...(((((((((.....-...)))))))))(--((((..-----((......)))))))... ( -26.60) >DroAna_CAF1 11935 113 + 1 UUAUUGUGCAUU-UUAUGCAUUUUGUGCGCGAUUUCGAU-UUGAAGUUGGGCUCUGAAAUAAAUAUGAAAAGUU-UUAUUUAUGUUUG--UGGGUUGGUGGCGGAAAAGCCAAGCAAA- .....((((((.-..))))))(((((((.(((((((...-..))))))).)).(..(.(((((((((((.....-...))))))))))--)...)..)((((......)))).)))))- ( -29.10) >consensus UUAUUGUGCAUU_UUAUGCAUUUUGUGCGCGAUUUCGAU_UUGAAGUUG_GCUCUGAAAUAAAUAUGAAAAGUU_UUAUUUAUGUUUG__CGGGC______GGGCAAAGCCAGGGAAAG .....((((((....)))))).....((.((((((((....)))))))).)).(((...((((((((((.........))))))))))..))).......................... (-18.74 = -17.88 + -0.86)


| Location | 22,093,618 – 22,093,714 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.35 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -15.32 |
| Energy contribution | -16.24 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.50 |
| SVM decision value | 3.70 |
| SVM RNA-class probability | 0.999538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22093618 96 + 27905053 UUGAAGUUGGGCUCUGAAAUAAAUAUGAAAAGUUUUAUUUAUGUUUGUGGGG------UGGCAAAGCCCGGAAAAAAGGGGCCCUGGCU-UUG----------GUUUGUGUUC (..((((((((((((...(((((((((((........))))))))))).(((------(......))))........))))))).))))-)..----------)......... ( -30.50) >DroEre_CAF1 16711 102 + 1 UUGAAGUUGGGCUCUGAAAUAAAUAUGAAAAGUUUUAUUUAUGUUUGUGGGG------UGGCAAAGCCAG-AAAAAAGGGGGCCUGGCU-UUGGAAC---UCAGUUUGUGUUC ..(((((..((((((...(((((((((((........)))))))))))....------((((...)))).-.......))))))..)))-)).((((---.(.....).)))) ( -30.80) >DroYak_CAF1 15669 103 + 1 UUGAAGUUGGGCUCUGAAAUAAAUAUGAAAAGUUUUAUUUAUGUUUGUGGGG------UGGCAAAGCCAGCAAAAAAGGGGGCCUGGCU-UUGGAGC---UCAGUUUGUGUUC ..(((((..((((((...(((((((((((........)))))))))))..(.------((((...)))).).......))))))..)))-)).((((---.(.....).)))) ( -33.00) >DroMoj_CAF1 83232 89 + 1 UCGAAGUUA-GCUUGGAAAUAAAUAUGAAAAGUUUUAUUUAUGUUUGCCACCC-----GGACUAAGCCGUGGCUAGA--GCGCCUAGCA-UCGA---------------GCGU ((((.((((-((((((...((((((((((........))))))))))((....-----)).))))))...(((....--..))))))).-))))---------------.... ( -23.00) >DroAna_CAF1 11973 111 + 1 UUGAAGUUGGGCUCUGAAAUAAAUAUGAAAAGUUUUAUUUAUGUUUGUGGGUUGGUGGCGGAAAAGCCAAGCAAA--UGGGGCGUGGCCGGUGGGUCUUUUCCAUUGGCUUUU ..........(((((...(((((((((((........)))))))))))...(((.((((......))))..))).--.)))))..((((((((((.....))))))))))... ( -34.50) >consensus UUGAAGUUGGGCUCUGAAAUAAAUAUGAAAAGUUUUAUUUAUGUUUGUGGGG______UGGCAAAGCCAGGAAAAAAGGGGGCCUGGCU_UUGG__C___UC_GUUUGUGUUC .....((((((((((...(((((((((((........)))))))))))...........((.....))..........))))))))))......................... (-15.32 = -16.24 + 0.92)



| Location | 22,093,618 – 22,093,714 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.35 |
| Mean single sequence MFE | -16.66 |
| Consensus MFE | -4.69 |
| Energy contribution | -5.13 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.28 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22093618 96 - 27905053 GAACACAAAC----------CAA-AGCCAGGGCCCCUUUUUUCCGGGCUUUGCCA------CCCCACAAACAUAAAUAAAACUUUUCAUAUUUAUUUCAGAGCCCAACUUCAA (((.......----------...-.((....))...........((((((((...------............(((((((..........)))))))))))))))...))).. ( -14.26) >DroEre_CAF1 16711 102 - 1 GAACACAAACUGA---GUUCCAA-AGCCAGGCCCCCUUUUUU-CUGGCUUUGCCA------CCCCACAAACAUAAAUAAAACUUUUCAUAUUUAUUUCAGAGCCCAACUUCAA .........((((---((..(((-(((((((..........)-)))))))))..)------).........(((((((..........))))))).))))............. ( -17.80) >DroYak_CAF1 15669 103 - 1 GAACACAAACUGA---GCUCCAA-AGCCAGGCCCCCUUUUUUGCUGGCUUUGCCA------CCCCACAAACAUAAAUAAAACUUUUCAUAUUUAUUUCAGAGCCCAACUUCAA ..........((.---(((((((-((((((.............)))))))))...------..........(((((((..........)))))))....)))).))....... ( -19.02) >DroMoj_CAF1 83232 89 - 1 ACGC---------------UCGA-UGCUAGGCGC--UCUAGCCACGGCUUAGUCC-----GGGUGGCAAACAUAAAUAAAACUUUUCAUAUUUAUUUCCAAGC-UAACUUCGA .(((---------------(((.-.(((((((((--....))....))))))).)-----)))))((....(((((((..........)))))))......))-......... ( -17.40) >DroAna_CAF1 11973 111 - 1 AAAAGCCAAUGGAAAAGACCCACCGGCCACGCCCCA--UUUGCUUGGCUUUUCCGCCACCAACCCACAAACAUAAAUAAAACUUUUCAUAUUUAUUUCAGAGCCCAACUUCAA ..((((.(((((....(......)(((...))))))--)).))))(((......)))..............(((((((..........))))))).................. ( -14.80) >consensus GAACACAAAC_GA___G__CCAA_AGCCAGGCCCCCUUUUUUCCUGGCUUUGCCA______CCCCACAAACAUAAAUAAAACUUUUCAUAUUUAUUUCAGAGCCCAACUUCAA .............................................(((((((.....................(((((((..........))))))))))))))......... ( -4.69 = -5.13 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:07:00 2006