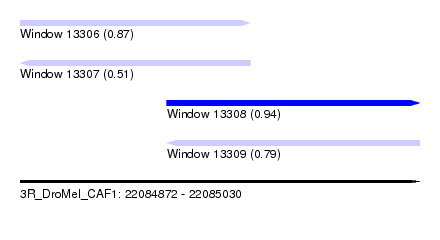
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,084,872 – 22,085,030 |
| Length | 158 |
| Max. P | 0.938242 |

| Location | 22,084,872 – 22,084,963 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 85.85 |
| Mean single sequence MFE | -37.28 |
| Consensus MFE | -30.59 |
| Energy contribution | -30.07 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22084872 91 + 27905053 CGCUGCUGCUG-CGCUGUUCCUGCAGGCAGCGACGCCAACGCAGGGCGCAUGAAUUUUCGCUUGACAUUGAACAAGGUCAAGCAACAAGC---CU .((((((((((-(.((((....))))))))))..(((.......)))))..........(((((((.((....)).)))))))....)))---.. ( -33.40) >DroSec_CAF1 7641 91 + 1 CGCUUCUGCUG-CGCUGCCCCUGCAGGCAGCGACGCUAACGCAGGGCGCAUGAAUUUUCGCUUGACAUUGAACAAGGUCAAGCAACAAGC---CU .((....((..-(((((((......)))))))..))....)).((((....((....))(((((((.((....)).))))))).....))---)) ( -34.20) >DroSim_CAF1 8081 91 + 1 CGCUGCUGCUG-CGCUGCCCCUGCAGGCAGCGACGCCAACGCAGGGCGCAUGAAUUUUCGCUUGACAUUGAACAAGGUCAAGCAACAAGC---CU ..((((.((..-(((((((......)))))))..))....))))(((....((....))(((((((.((....)).))))))).....))---). ( -35.50) >DroEre_CAF1 7738 91 + 1 CGCUGCUGCUG-CGCUGCCCCUGCAGGCAGCGACGGCAACGCAGGGCGCAUGAAUUUUCGCCUGACAUUGAACAAGGUCAAGCAACAAGC---CU ..(((((((((-(((((((......))))))).)))))..))))(((....((....))((.((((.((....)).)))).)).....))---). ( -37.10) >DroYak_CAF1 6596 91 + 1 CGCUGCUGCUG-CUCUGCCGCUGCAGGCAGCGACGGCAACGCAGGGCGCAUGUAUUUUCGCUUGACAUUGAACAAGGUCAAGCAACAAGC---CU .((((((((.(-(((((((((((....)))))..(....))))))))))).))......(((((((.((....)).)))))))....)))---.. ( -38.90) >DroAna_CAF1 3705 95 + 1 CGCUGCCAACGUUGCUGCCGACGACGGCAGAGAUGCGGGCGUGGGGCGCAUGUGUUGGGGCCUGACAUUGAACAAGGUCAGGCAACAAUGUUGCC (((.(((..(((..((((((....))))))..)))..)))))).((((...(..(((..(((((((.((....)).)))))))..)))..))))) ( -44.60) >consensus CGCUGCUGCUG_CGCUGCCCCUGCAGGCAGCGACGCCAACGCAGGGCGCAUGAAUUUUCGCUUGACAUUGAACAAGGUCAAGCAACAAGC___CU ((((.((((...(((((((......)))))))..(....)))))))))...........(((((((.((....)).)))))))............ (-30.59 = -30.07 + -0.52)


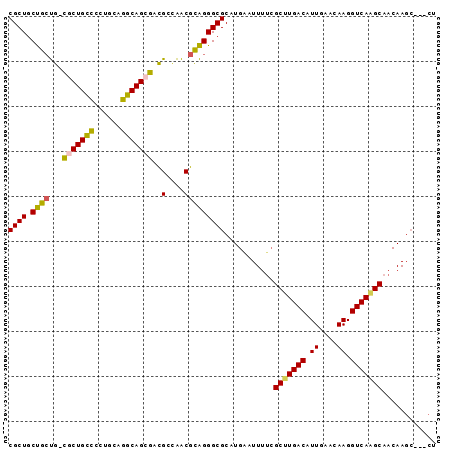
| Location | 22,084,872 – 22,084,963 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 85.85 |
| Mean single sequence MFE | -37.15 |
| Consensus MFE | -27.85 |
| Energy contribution | -28.22 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22084872 91 - 27905053 AG---GCUUGUUGCUUGACCUUGUUCAAUGUCAAGCGAAAAUUCAUGCGCCCUGCGUUGGCGUCGCUGCCUGCAGGAACAGCG-CAGCAGCAGCG ..---((...(((((((((.((....)).))))))))).......(((...((((((((...((.(((....))))).)))))-)))..))))). ( -35.40) >DroSec_CAF1 7641 91 - 1 AG---GCUUGUUGCUUGACCUUGUUCAAUGUCAAGCGAAAAUUCAUGCGCCCUGCGUUAGCGUCGCUGCCUGCAGGGGCAGCG-CAGCAGAAGCG ..---((((.(((((((((.((....)).))))))))).......(((.....((....))(.((((((((....))))))))-).))).)))). ( -36.00) >DroSim_CAF1 8081 91 - 1 AG---GCUUGUUGCUUGACCUUGUUCAAUGUCAAGCGAAAAUUCAUGCGCCCUGCGUUGGCGUCGCUGCCUGCAGGGGCAGCG-CAGCAGCAGCG ..---(((.((((((((((.((....)).)))))(((........(((.(((((((..((((....)))))))))))))).))-).)))))))). ( -38.20) >DroEre_CAF1 7738 91 - 1 AG---GCUUGUUGCUUGACCUUGUUCAAUGUCAGGCGAAAAUUCAUGCGCCCUGCGUUGCCGUCGCUGCCUGCAGGGGCAGCG-CAGCAGCAGCG ..---((...(((((((((.((....)).))))))))).......(((...((((((((((.(.((.....)).).)))))))-)))..))))). ( -39.80) >DroYak_CAF1 6596 91 - 1 AG---GCUUGUUGCUUGACCUUGUUCAAUGUCAAGCGAAAAUACAUGCGCCCUGCGUUGCCGUCGCUGCCUGCAGCGGCAGAG-CAGCAGCAGCG .(---((.(((((((((((.((....)).)))))))).....)))...)))(((((((((((((((((....))))))).).)-)))).)))).. ( -38.60) >DroAna_CAF1 3705 95 - 1 GGCAACAUUGUUGCCUGACCUUGUUCAAUGUCAGGCCCCAACACAUGCGCCCCACGCCCGCAUCUCUGCCGUCGUCGGCAGCAACGUUGGCAGCG (((..((((((((((((((.((....)).)))))))...)))).))).)))....(((.((....((((((....))))))....)).))).... ( -34.90) >consensus AG___GCUUGUUGCUUGACCUUGUUCAAUGUCAAGCGAAAAUUCAUGCGCCCUGCGUUGGCGUCGCUGCCUGCAGGGGCAGCG_CAGCAGCAGCG ..........(((((((((.((....)).)))))))))........((...((((........((((((((....))))))))...))))..)). (-27.85 = -28.22 + 0.36)


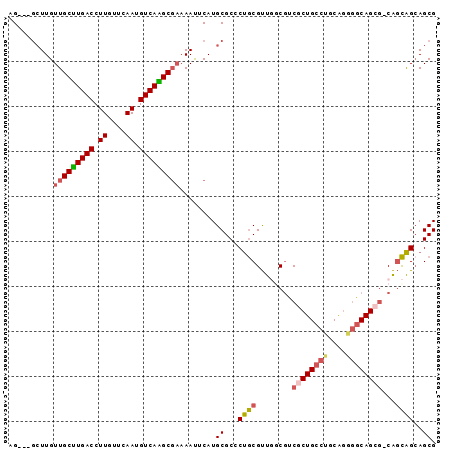
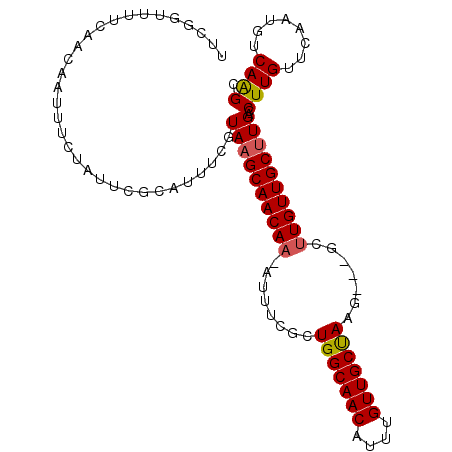
| Location | 22,084,930 – 22,085,030 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 92.57 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -21.81 |
| Energy contribution | -21.97 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22084930 100 + 27905053 GCUUGACAUUGAACAAGGUCAAGCAACAAGC---CUUAGCAACAAAUGUUGCCAGCGAAAU-UUGUUGCUUACGAAAUGCGAAUAGAAAUUGUUGAAAACCGAA (((((((.((....)).)))))))(((((..---...((((((((((.(((....))).))-))))))))..((.....))........))))).......... ( -25.80) >DroSec_CAF1 7699 100 + 1 GCUUGACAUUGAACAAGGUCAAGCAACAAGC---CUUAGCAACAAAUGUUGCCAGCGAAAU-UUGUUGCUUACGAAAUGCGAAUAGAAAUUGUUGAAAACCGAA (((((((.((....)).)))))))(((((..---...((((((((((.(((....))).))-))))))))..((.....))........))))).......... ( -25.80) >DroSim_CAF1 8139 100 + 1 GCUUGACAUUGAACAAGGUCAAGCAACAAGC---CUUAGCAACAAAUGUUGCCAGCGAAAU-UUGUUGCUUACGAAAUGCGAAUAGAAAUUGUUGAAAACCGAA (((((((.((....)).)))))))(((((..---...((((((((((.(((....))).))-))))))))..((.....))........))))).......... ( -25.80) >DroEre_CAF1 7796 100 + 1 GCCUGACAUUGAACAAGGUCAAGCAACAAGC---CUUAGCAACAAAUGUUGCCAGCGAAAU-UUGUUGCUUACGAAAUGCGAAUAGAAAUUGUUGAAAACCGAA ........((.(((((..(((((((((((((---....(((((....)))))..)).....-))))))))).((.....))....))..))))).))....... ( -22.90) >DroYak_CAF1 6654 100 + 1 GCUUGACAUUGAACAAGGUCAAGCAACAAGC---CUUAGCAACAAAUGUUGCCAGCGAAAU-UUGUUGCUUACGAAAUGCGAAUAGAAAUUGUUGAAAACCGAA (((((((.((....)).)))))))(((((..---...((((((((((.(((....))).))-))))))))..((.....))........))))).......... ( -25.80) >DroAna_CAF1 3764 102 + 1 GCCUGACAUUGAACAAGGUCAGGCAACAAUGUUGCCUGGCAAC--AUGUUGCCAGCAAGACAAUGUUGCGUACGAAACCCACAUAGAAAUUGUCGAAAACCGAA (((((((.((....)).)))))))((((.((((..(((((((.--...)))))))...)))).)))).........................(((.....))). ( -31.30) >consensus GCUUGACAUUGAACAAGGUCAAGCAACAAGC___CUUAGCAACAAAUGUUGCCAGCGAAAU_UUGUUGCUUACGAAAUGCGAAUAGAAAUUGUUGAAAACCGAA (((((((.((....)).)))))))..............(((((....)))))((((((.......((((.........)))).......))))))......... (-21.81 = -21.97 + 0.17)

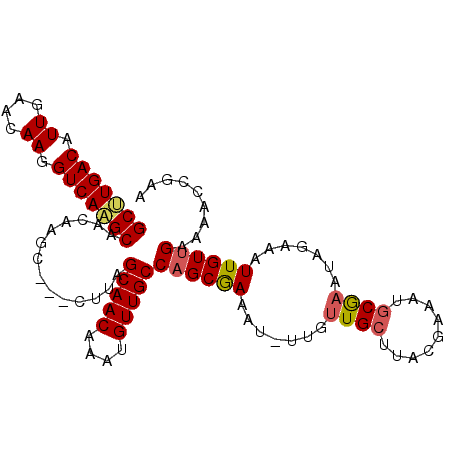

| Location | 22,084,930 – 22,085,030 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 92.57 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -20.92 |
| Energy contribution | -20.89 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22084930 100 - 27905053 UUCGGUUUUCAACAAUUUCUAUUCGCAUUUCGUAAGCAACAA-AUUUCGCUGGCAACAUUUGUUGCUAAG---GCUUGUUGCUUGACCUUGUUCAAUGUCAAGC ...((..((.(((((..((.....((.....))(((((((((-.....(((((((((....))))))..)---)))))))))))))..))))).))..)).... ( -22.50) >DroSec_CAF1 7699 100 - 1 UUCGGUUUUCAACAAUUUCUAUUCGCAUUUCGUAAGCAACAA-AUUUCGCUGGCAACAUUUGUUGCUAAG---GCUUGUUGCUUGACCUUGUUCAAUGUCAAGC ...((..((.(((((..((.....((.....))(((((((((-.....(((((((((....))))))..)---)))))))))))))..))))).))..)).... ( -22.50) >DroSim_CAF1 8139 100 - 1 UUCGGUUUUCAACAAUUUCUAUUCGCAUUUCGUAAGCAACAA-AUUUCGCUGGCAACAUUUGUUGCUAAG---GCUUGUUGCUUGACCUUGUUCAAUGUCAAGC ...((..((.(((((..((.....((.....))(((((((((-.....(((((((((....))))))..)---)))))))))))))..))))).))..)).... ( -22.50) >DroEre_CAF1 7796 100 - 1 UUCGGUUUUCAACAAUUUCUAUUCGCAUUUCGUAAGCAACAA-AUUUCGCUGGCAACAUUUGUUGCUAAG---GCUUGUUGCUUGACCUUGUUCAAUGUCAGGC ...((..((.(((((..((.....((.....))(((((((((-.....(((((((((....))))))..)---)))))))))))))..))))).))..)).... ( -22.50) >DroYak_CAF1 6654 100 - 1 UUCGGUUUUCAACAAUUUCUAUUCGCAUUUCGUAAGCAACAA-AUUUCGCUGGCAACAUUUGUUGCUAAG---GCUUGUUGCUUGACCUUGUUCAAUGUCAAGC ...((..((.(((((..((.....((.....))(((((((((-.....(((((((((....))))))..)---)))))))))))))..))))).))..)).... ( -22.50) >DroAna_CAF1 3764 102 - 1 UUCGGUUUUCGACAAUUUCUAUGUGGGUUUCGUACGCAACAUUGUCUUGCUGGCAACAU--GUUGCCAGGCAACAUUGUUGCCUGACCUUGUUCAAUGUCAGGC ..(((...((.(((.......))).))..)))...((((((.(((..(((((((((...--.)))))).)))))).))))))(((((.((....)).))))).. ( -31.90) >consensus UUCGGUUUUCAACAAUUUCUAUUCGCAUUUCGUAAGCAACAA_AUUUCGCUGGCAACAUUUGUUGCUAAG___GCUUGUUGCUUGACCUUGUUCAAUGUCAAGC ................................((((((((((........(((((((....))))))).......))))))))))..((((........)))). (-20.92 = -20.89 + -0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:55 2006