
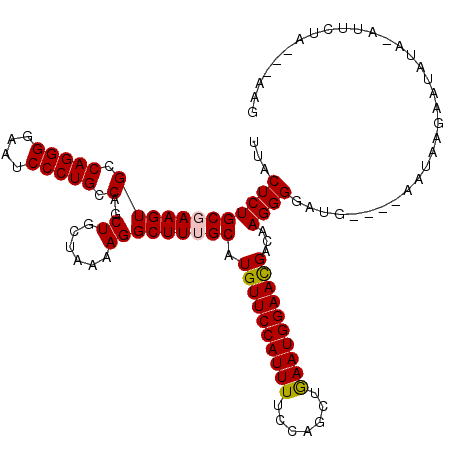
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,075,724 – 22,075,933 |
| Length | 209 |
| Max. P | 0.932614 |

| Location | 22,075,724 – 22,075,833 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 89.20 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -29.28 |
| Energy contribution | -29.36 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22075724 109 + 27905053 UUACUCUGCGAAGUGCCAGGGGAAUCCCUGCCAGCUGCUAAAAGGCUUUGCAUGUUCCAUUUUCCAGCUGAAUGGAACGGCAAGGGGAUG----AAUAAGAAUAU--AUUCUA----AG ...(((((.(.....))))))..(((((((((.((.(((....)))...))..(((((((((.......))))))))))))..))))))(----((((......)--))))..----.. ( -35.30) >DroSec_CAF1 67773 115 + 1 UUACUCUGCCAAGUGCCAGGGGAAUCCCUGCCAGCUGCUAAAAGGCUUUGCAUGUUCCAUUUUCCAGCUGAAUGGAACGACAAGGGGAUG----AAUAAGAAUAUAUAUUCUAAGUAAG ...(((((.(....).)))))..((((((((.((((.......))))..)).((((((((((.......))))))))))....)))))).----....(((((....)))))....... ( -31.60) >DroSim_CAF1 61213 115 + 1 UUACUCUGCCAAGUGCCAGGGGAAUCCCUGCCAGCUGCUAAAAGGCUUUGCAUGUUCCAUUUUCCAGCUGAAUGGAACGACAAGGGGAUG----AAUAAGAAUAUAUAUUCUAAGUAAG ...(((((.(....).)))))..((((((((.((((.......))))..)).((((((((((.......))))))))))....)))))).----....(((((....)))))....... ( -31.60) >DroEre_CAF1 62403 105 + 1 UUACUCUGCGAAGUGCCAGGGGAAUCCCUGCCAGCUGCUAAAAGGCUUUGCGUGUUCCAUUUUCCGGCUAAAUGGAAUGGCAAGGGGAUG----CAUAAGAAUAUA----------AAG ...(((((.(.....))))))(.(((((((((.((.(((....)))...))..(((((((((.......))))))))))))..)))))).----)...........----------... ( -30.80) >DroYak_CAF1 70253 109 + 1 UUACUCUGCGAAGUGCCAGGGGAAUCCCUGCCAGCUGCUAAAAGGCUUUGCAUGUUCCAUUUUCCAGCUGAAUGGAACGCCAAGGGGAUGGAUCCGUAAGAAUAUA----------AAG ...((((((((((((.(((((....))))).)..((......)))))))))..(((((((((.......)))))))))....)))).(((....))).........----------... ( -31.50) >consensus UUACUCUGCGAAGUGCCAGGGGAAUCCCUGCCAGCUGCUAAAAGGCUUUGCAUGUUCCAUUUUCCAGCUGAAUGGAACGACAAGGGGAUG____AAUAAGAAUAUA_AUUCUA___AAG ...((((((((((((.(((((....))))).)..((......))))))))).((((((((((.......))))))))))...))))................................. (-29.28 = -29.36 + 0.08)

| Location | 22,075,724 – 22,075,833 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.20 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -25.92 |
| Energy contribution | -26.12 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22075724 109 - 27905053 CU----UAGAAU--AUAUUCUUAUU----CAUCCCCUUGCCGUUCCAUUCAGCUGGAAAAUGGAACAUGCAAAGCCUUUUAGCAGCUGGCAGGGAUUCCCCUGGCACUUCGCAGAGUAA ..----..((((--(......))))----).......((((((((((((.........)))))))).(((...((......))((.((.(((((....))))).))))..)))).))). ( -28.50) >DroSec_CAF1 67773 115 - 1 CUUACUUAGAAUAUAUAUUCUUAUU----CAUCCCCUUGUCGUUCCAUUCAGCUGGAAAAUGGAACAUGCAAAGCCUUUUAGCAGCUGGCAGGGAUUCCCCUGGCACUUGGCAGAGUAA ..(((((.(((((........))))----).......((((((((((((.........)))))))).(((...((......))......(((((....))))))))...))))))))). ( -30.80) >DroSim_CAF1 61213 115 - 1 CUUACUUAGAAUAUAUAUUCUUAUU----CAUCCCCUUGUCGUUCCAUUCAGCUGGAAAAUGGAACAUGCAAAGCCUUUUAGCAGCUGGCAGGGAUUCCCCUGGCACUUGGCAGAGUAA ..(((((.(((((........))))----).......((((((((((((.........)))))))).(((...((......))......(((((....))))))))...))))))))). ( -30.80) >DroEre_CAF1 62403 105 - 1 CUU----------UAUAUUCUUAUG----CAUCCCCUUGCCAUUCCAUUUAGCCGGAAAAUGGAACACGCAAAGCCUUUUAGCAGCUGGCAGGGAUUCCCCUGGCACUUCGCAGAGUAA ...----------..((((((...(----(......((((..((((((((.......))))))))...)))).........(.((.((.(((((....))))).)))).))))))))). ( -27.70) >DroYak_CAF1 70253 109 - 1 CUU----------UAUAUUCUUACGGAUCCAUCCCCUUGGCGUUCCAUUCAGCUGGAAAAUGGAACAUGCAAAGCCUUUUAGCAGCUGGCAGGGAUUCCCCUGGCACUUCGCAGAGUAA ...----------..((((((..((((...........(((((((((((.........)))))))).(((...........))))))(.(((((....))))).)..)))).)))))). ( -29.30) >consensus CUU___UAGAAU_UAUAUUCUUAUU____CAUCCCCUUGCCGUUCCAUUCAGCUGGAAAAUGGAACAUGCAAAGCCUUUUAGCAGCUGGCAGGGAUUCCCCUGGCACUUCGCAGAGUAA ...............((((((....................((((((((.........))))))))..((...((......))((.((.(((((....))))).))))..)))))))). (-25.92 = -26.12 + 0.20)


| Location | 22,075,763 – 22,075,873 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.43 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -22.90 |
| Energy contribution | -22.58 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22075763 110 + 27905053 AAAAGGCUUUGCAUGUUCCAUUUUCCAGCUGAAUGGAACGGCAAGGGGAUG----AAUAAGAAUAU--AUUCUA----AGUAUAAUAUGUUCCCUCUCAUUACACACACUAGCGGCAGCA ......((((((.((((((((((.......)))))))))))))))).((((----(....((((((--(((...----.....)))))))))....)))))..........((....)). ( -30.30) >DroSec_CAF1 67812 114 + 1 AAAAGGCUUUGCAUGUUCCAUUUUCCAGCUGAAUGGAACGACAAGGGGAUG----AAUAAGAAUAUAUAUUCUAAGUAAGUAUAAUAAGUUCCCUCUCAUUACAC--ACUCGCGGCAGCA .....(((((((..(((((((((.......)))))))))((..(((((((.----.......((((.(((.....))).)))).....))))))).)).......--....)))).))). ( -27.02) >DroSim_CAF1 61252 114 + 1 AAAAGGCUUUGCAUGUUCCAUUUUCCAGCUGAAUGGAACGACAAGGGGAUG----AAUAAGAAUAUAUAUUCUAAGUAAGUAUAAUAAGUUCCCUCUCAUUACAC--ACUCGCGGCAGCA .....(((((((..(((((((((.......)))))))))((..(((((((.----.......((((.(((.....))).)))).....))))))).)).......--....)))).))). ( -27.02) >DroEre_CAF1 62442 104 + 1 AAAAGGCUUUGCGUGUUCCAUUUUCCGGCUAAAUGGAAUGGCAAGGGGAUG----CAUAAGAAUAUA----------AAGUAUAAUAAGUUCCCCCUCAUUACAC--ACUCGCGGCAGCC ....(((((((((.(((((((((.......)))))))))....(((((..(----(...........----------...........))..)))))........--...))))).)))) ( -26.15) >DroYak_CAF1 70292 108 + 1 AAAAGGCUUUGCAUGUUCCAUUUUCCAGCUGAAUGGAACGCCAAGGGGAUGGAUCCGUAAGAAUAUA----------AAGUAUAAUAUGUUCCCUCUCAUUACAC--ACUCGCGGCAGCA .....(((((((..(((((((((.......)))))))))....((((((((....)))..(((((((----------........))))))))))))........--....)))).))). ( -28.40) >consensus AAAAGGCUUUGCAUGUUCCAUUUUCCAGCUGAAUGGAACGACAAGGGGAUG____AAUAAGAAUAUA_AUUCUA___AAGUAUAAUAAGUUCCCUCUCAUUACAC__ACUCGCGGCAGCA .....(((((((.((((((((((.......))))))))))....((((((......................................)))))).................)))).))). (-22.90 = -22.58 + -0.32)



| Location | 22,075,803 – 22,075,913 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.61 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -26.86 |
| Energy contribution | -27.42 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22075803 110 - 27905053 ACCAUUUAAUUCAAAUGGGUGGGCCGCAGAGCAAACGUUUUGCUGCCGCUAGUGUGUGUAAUGAGAGGGAACAUAUUAUACU----UAGAAU--AUAUUCUUAUU----CAUCCCCUUGC .((((((.....))))))...(((.(((((((....))))))).)))((.((.(..((.((((((((....)(((((.....----...)))--))..)))))))----))..).)).)) ( -29.40) >DroSec_CAF1 67852 114 - 1 ACCAUUUAAUUCAAAUGGGUGGGCCGCAGAGCAAACGUUUUGCUGCCGCGAGU--GUGUAAUGAGAGGGAACUUAUUAUACUUACUUAGAAUAUAUAUUCUUAUU----CAUCCCCUUGU .((((((.....))))))...(((.(((((((....))))))).)))(((((.--((((((((((......)))))))))).......(((((........))))----).....))))) ( -34.90) >DroSim_CAF1 61292 114 - 1 ACCAUUUAAUUCAAAUGGGUGGGCCGCAGAGCAAACGUUUUGCUGCCGCGAGU--GUGUAAUGAGAGGGAACUUAUUAUACUUACUUAGAAUAUAUAUUCUUAUU----CAUCCCCUUGU .((((((.....))))))...(((.(((((((....))))))).)))(((((.--((((((((((......)))))))))).......(((((........))))----).....))))) ( -34.90) >DroEre_CAF1 62482 104 - 1 AGCAUUUAAUUCGAAUGGGUGGACCGCAGAGCAAACGUUUGGCUGCCGCGAGU--GUGUAAUGAGGGGGAACUUAUUAUACUU----------UAUAUUCUUAUG----CAUCCCCUUGC .((((...(((((....((..(.((...((((....)))))))..)).)))))--((((((((((......))))))))))..----------.........)))----).......... ( -26.50) >DroYak_CAF1 70332 108 - 1 ACCAUUUAAUUCGAAUGGGUGGGCCGCAGAGCAAACGUUUUGCUGCCGCGAGU--GUGUAAUGAGAGGGAACAUAUUAUACUU----------UAUAUUCUUACGGAUCCAUCCCCUUGG .((((((.....))))))...(((.(((((((....))))))).))).((((.--((((((((...(....).))))))))..----------...........(((....))).)))). ( -28.70) >consensus ACCAUUUAAUUCAAAUGGGUGGGCCGCAGAGCAAACGUUUUGCUGCCGCGAGU__GUGUAAUGAGAGGGAACUUAUUAUACUU___UAGAAU_UAUAUUCUUAUU____CAUCCCCUUGC ...........(((..((((((((.(((((((....))))))).)))........((((((((((......))))))))))............................)))))..))). (-26.86 = -27.42 + 0.56)


| Location | 22,075,833 – 22,075,933 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 95.95 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -23.04 |
| Energy contribution | -23.60 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22075833 100 - 27905053 AAUGAGCAAGUGCAAUGAGUACCAUUUAAUUCAAAUGGGUGGGCCGCAGAGCAAACGUUUUGCUGCCGCUAGUGUGUGUAAUGAGAGGGAACAUAUUAUA ....(((..............((((((.....))))))...(((.(((((((....))))))).))))))(((((((....(....)...)))))))... ( -27.60) >DroSec_CAF1 67888 98 - 1 AAUGAGCAAGUGCAAUGAGUACCAUUUAAUUCAAAUGGGUGGGCCGCAGAGCAAACGUUUUGCUGCCGCGAGU--GUGUAAUGAGAGGGAACUUAUUAUA .....(((..(((........((((((.....))))))...(((.(((((((....))))))).))))))..)--))((((((((......)))))))). ( -31.40) >DroSim_CAF1 61328 98 - 1 AAUGAGCAAGUGCAAUGAGUACCAUUUAAUUCAAAUGGGUGGGCCGCAGAGCAAACGUUUUGCUGCCGCGAGU--GUGUAAUGAGAGGGAACUUAUUAUA .....(((..(((........((((((.....))))))...(((.(((((((....))))))).))))))..)--))((((((((......)))))))). ( -31.40) >DroEre_CAF1 62508 98 - 1 AAUGAGCAAGUGCAAUGAGUAGCAUUUAAUUCGAAUGGGUGGACCGCAGAGCAAACGUUUGGCUGCCGCGAGU--GUGUAAUGAGGGGGAACUUAUUAUA .....(((..(((.........(((((.....)))))((..(.((...((((....)))))))..)))))..)--))((((((((......)))))))). ( -24.00) >DroYak_CAF1 70362 98 - 1 AAUGAGCAAGUGCAAUGAGUACCAUUUAAUUCGAAUGGGUGGGCCGCAGAGCAAACGUUUUGCUGCCGCGAGU--GUGUAAUGAGAGGGAACAUAUUAUA .....(((..(((........((((((.....))))))...(((.(((((((....))))))).))))))..)--))((((((...(....).)))))). ( -27.60) >consensus AAUGAGCAAGUGCAAUGAGUACCAUUUAAUUCAAAUGGGUGGGCCGCAGAGCAAACGUUUUGCUGCCGCGAGU__GUGUAAUGAGAGGGAACUUAUUAUA .....((....))........((((((.....))))))((((.(.(((((((....))))))).))))).......(((((((((......))))))))) (-23.04 = -23.60 + 0.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:50 2006