
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,070,456 – 22,070,569 |
| Length | 113 |
| Max. P | 0.993867 |

| Location | 22,070,456 – 22,070,569 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.26 |
| Mean single sequence MFE | -26.94 |
| Consensus MFE | -24.47 |
| Energy contribution | -24.87 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22070456 113 + 27905053 GACCAUU-UACUUUUUAUUUCCAGAUGUGUCGCAAGAUUUGCUUCCGCAUUUUACUCAUUGCAAUGCAAAAGAGGUGCCA--CAUCUCCAUUGCAGUC----GGCCGCAAAUGAAAAUAA (((((((-(.............))))).))).....((((((..(((..........(((((((((.....((((((...--))))))))))))))))----))..))))))........ ( -26.02) >DroSec_CAF1 62504 116 + 1 GACCAUUUUACUCUUUAUUUCCAGAUGUGUCGCAAGAUUUGCUUCCGCAUUUUACUCAUUGCAAUGCAAAAGAGGUGCCACACAUCUCCAUUGCAUUC----GGCCGCAAAUGAAAAUAA ((((((((..............))))).))).....((((((..(((((..........)))(((((((..((((((.....))))))..))))))).----))..))))))........ ( -28.24) >DroSim_CAF1 55492 114 + 1 GACCAUUUUACUCUUUAUUUCCAGAUGUGUCGCAAGAUUUGCUUCCGCAUUUUACUCAUUGCAAUGCAAAAGAGGUGCCA--CAUCUCCAUUGCAGUC----GGCCGCAAAUGAAAAUAA ((((((((..............))))).))).....((((((..(((..........(((((((((.....((((((...--))))))))))))))))----))..))))))........ ( -25.74) >DroEre_CAF1 57008 112 + 1 GACCAUU-UACUUUUUAUUUCCAGAUGUGUCGCAAGAU-AGCUUCCGCAUUUUACUCAUUGCAAUGCAAAAGAGGUGCCA--CAUCUCCAUUGCACUC----GGCCGCAAAUGAAAAUAA .......-...(((((((((.......((((....)))-)((..(((((..........)))..(((((..((((((...--))))))..)))))...----))..)))))))))))... ( -25.90) >DroYak_CAF1 64946 117 + 1 GACCAUU-UACUUUUUAUUUCCAGAUGUGUCGCAAGAUUUGCUUCCGCAUUUUACUCAUUGCAAUGCAAAAGAGGUGCCA--CAUCUCUAUUGCAGUCGGUCGGCCGCAAAUGAAAAUAA .......-......((((((.((..((((((....))...(((.(((((..........)))..(((((.(((((((...--))))))).)))))...))..)))))))..)).)))))) ( -28.80) >consensus GACCAUU_UACUUUUUAUUUCCAGAUGUGUCGCAAGAUUUGCUUCCGCAUUUUACUCAUUGCAAUGCAAAAGAGGUGCCA__CAUCUCCAUUGCAGUC____GGCCGCAAAUGAAAAUAA (((((((................)))).))).....((((((..(((((..........)))..(((((..((((((.....))))))..))))).......))..))))))........ (-24.47 = -24.87 + 0.40)

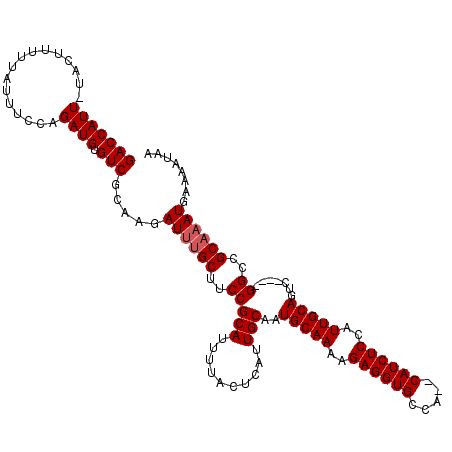

| Location | 22,070,456 – 22,070,569 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.26 |
| Mean single sequence MFE | -32.74 |
| Consensus MFE | -28.24 |
| Energy contribution | -28.28 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22070456 113 - 27905053 UUAUUUUCAUUUGCGGCC----GACUGCAAUGGAGAUG--UGGCACCUCUUUUGCAUUGCAAUGAGUAAAAUGCGGAAGCAAAUCUUGCGACACAUCUGGAAAUAAAAAGUA-AAUGGUC ((((((((..((((((..----..))))))...(((((--(((((.......))).((((((.((......(((....)))..)))))))))))))))))))))))......-....... ( -32.90) >DroSec_CAF1 62504 116 - 1 UUAUUUUCAUUUGCGGCC----GAAUGCAAUGGAGAUGUGUGGCACCUCUUUUGCAUUGCAAUGAGUAAAAUGCGGAAGCAAAUCUUGCGACACAUCUGGAAAUAAAGAGUAAAAUGGUC ((((((((..(((((...----...)))))...((((((((.(((.(((..((((...)))).))).....(((....))).....))).)))))))))))))))).............. ( -35.60) >DroSim_CAF1 55492 114 - 1 UUAUUUUCAUUUGCGGCC----GACUGCAAUGGAGAUG--UGGCACCUCUUUUGCAUUGCAAUGAGUAAAAUGCGGAAGCAAAUCUUGCGACACAUCUGGAAAUAAAGAGUAAAAUGGUC ((((((((..((((((..----..))))))...(((((--(((((.......))).((((((.((......(((....)))..))))))))))))))))))))))).............. ( -32.90) >DroEre_CAF1 57008 112 - 1 UUAUUUUCAUUUGCGGCC----GAGUGCAAUGGAGAUG--UGGCACCUCUUUUGCAUUGCAAUGAGUAAAAUGCGGAAGCU-AUCUUGCGACACAUCUGGAAAUAAAAAGUA-AAUGGUC ((((((((..(((((...----...)))))...(((((--(((((.......))).((((((.((.......((....)).-.)))))))))))))))))))))))......-....... ( -28.90) >DroYak_CAF1 64946 117 - 1 UUAUUUUCAUUUGCGGCCGACCGACUGCAAUAGAGAUG--UGGCACCUCUUUUGCAUUGCAAUGAGUAAAAUGCGGAAGCAAAUCUUGCGACACAUCUGGAAAUAAAAAGUA-AAUGGUC ((((((((..((((((.(....).))))))...(((((--(((((.......))).((((((.((......(((....)))..)))))))))))))))))))))))......-....... ( -33.40) >consensus UUAUUUUCAUUUGCGGCC____GACUGCAAUGGAGAUG__UGGCACCUCUUUUGCAUUGCAAUGAGUAAAAUGCGGAAGCAAAUCUUGCGACACAUCUGGAAAUAAAAAGUA_AAUGGUC (((((((((..((.(..........(((((.((((.((.....)).)))).)))))((((((.((......(((....)))..))))))))).))..))))))))).............. (-28.24 = -28.28 + 0.04)

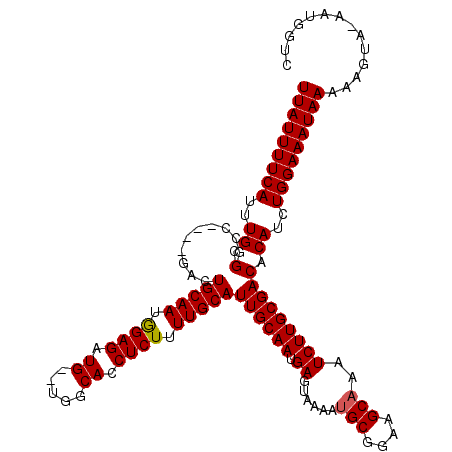
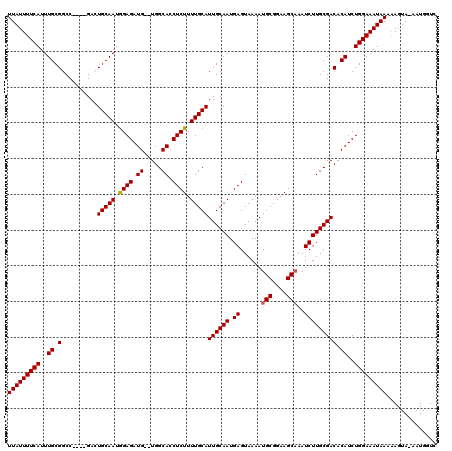
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:33 2006