
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,066,019 – 22,066,495 |
| Length | 476 |
| Max. P | 0.999482 |

| Location | 22,066,019 – 22,066,137 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.28 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -27.18 |
| Energy contribution | -27.78 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22066019 118 - 27905053 UUUUCAAUAGCAAGGACGUCAGCUGCCGGGCCUGAAAUGUUUAAUAGUGGGCUCGGCGGGUAAAGAGGAUUGUUUAAGGAAAAGUAGUUUCAUGGAUAUUUCCAUUUUCAUUUUU--UCG ........(((((...(.(((.((((((((((((..((.....))..)))))))))))).)...)).).)))))...((((((((.(....(((((....)))))...)))))))--)). ( -33.80) >DroSec_CAF1 58004 118 - 1 UUUUCAAUAGCAAGGACGUCAGCUGCCGGGCCUGAAAUGUUUAAUAGUGGGCUCGGCGGGUAAAGAGGAUUGGGUAAGGAAAAGUAAUUCCAUGGAUAUUUCCAUUUUAAUUUUU--UCG ......................((((((((((((..((.....))..))))))))))))....(((((((((((...((((......))))(((((....)))))))))))))))--).. ( -33.20) >DroSim_CAF1 51023 118 - 1 UUUUCAAUAGCAAGGACGUCAGCUGCCGGGCCUGAAAUGUUUAAUAGUGGGCUCGGCGGGUAAAGAGGAUCGGGUAAGGAAAAGUAGUUUCAUGGAUAUUUCCAUUUUAAUUUUU--UCG .........((..((.(.(((.((((((((((((..((.....))..)))))))))))).)...)).).))..))..((((((((......(((((....)))))....))))))--)). ( -33.80) >DroEre_CAF1 52556 118 - 1 UUUUCAAUAGCAAGGACGUCAGCUGCCGGGCCUGAAAUGUUUAAUAGUGGGCUGGGCGGGUAAAGAGGAUUGGUUAGGGAAAAGUAGUUUUAUGGAUAUUUCCAUUUUCCUUUUG--UCG ..............(((.....(((((.((((((..((.....))..)))))).)))))...((((((((((.((......)).)))....(((((....)))))..))))))))--)). ( -31.50) >DroYak_CAF1 60423 120 - 1 UUUUCAAUAGCAAGGACGUCAGCUGCCGGGCCUGAAAUGUUUAAUAGUGGGCUCGGCGGGUAAAGAGGAUUGGGUAAGGAAAAGUAGUUUCUUGGAUAUUUCCAUUUUCAUUUUUUUUUU ......................((((((((((((..((.....))..))))))))))))..((((((((.((((..((((((.....))))))(((....)))...)))).)))))))). ( -31.10) >consensus UUUUCAAUAGCAAGGACGUCAGCUGCCGGGCCUGAAAUGUUUAAUAGUGGGCUCGGCGGGUAAAGAGGAUUGGGUAAGGAAAAGUAGUUUCAUGGAUAUUUCCAUUUUCAUUUUU__UCG (((((..((.(.....(.(((.((((((((((((..((.....))..)))))))))))).)...)).)....).))..)))))........(((((....)))))............... (-27.18 = -27.78 + 0.60)



| Location | 22,066,057 – 22,066,177 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -42.18 |
| Consensus MFE | -39.12 |
| Energy contribution | -39.80 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999482 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22066057 120 - 27905053 ACUUUCCGGUUUUCUGGCGUGAACGGCAAAAACCCAGUAAUUUUCAAUAGCAAGGACGUCAGCUGCCGGGCCUGAAAUGUUUAAUAGUGGGCUCGGCGGGUAAAGAGGAUUGUUUAAGGA ....(((((((((.((.((....)).))))))))...((((((((....((......))...((((((((((((..((.....))..)))))))))))).....)))))))).....))) ( -39.00) >DroSec_CAF1 58042 120 - 1 ACUUUCCGGUUUUCUGGCGUGAACGGCAAAAACCCAGUAAUUUUCAAUAGCAAGGACGUCAGCUGCCGGGCCUGAAAUGUUUAAUAGUGGGCUCGGCGGGUAAAGAGGAUUGGGUAAGGA .(((((((((((((((.((....)).)....((((.............(((..........)))((((((((((..((.....))..))))))))))))))..))))))))))).))).. ( -43.10) >DroSim_CAF1 51061 120 - 1 ACUUUCCGGUUUUCUGGCGUGAACGGCAAAAACCCAGUAAUUUUCAAUAGCAAGGACGUCAGCUGCCGGGCCUGAAAUGUUUAAUAGUGGGCUCGGCGGGUAAAGAGGAUCGGGUAAGGA .(((((((((((((((.((....)).)....((((.............(((..........)))((((((((((..((.....))..))))))))))))))..))))))))))).))).. ( -45.10) >DroEre_CAF1 52594 120 - 1 ACUUUCCGGUUUUCUGGCGUGAACGGGAAAAACCCAGUAAUUUUCAAUAGCAAGGACGUCAGCUGCCGGGCCUGAAAUGUUUAAUAGUGGGCUGGGCGGGUAAAGAGGAUUGGUUAGGGA .((..(((((((((((((((....(((.....))).((.((.....)).))....)))))..(((((.((((((..((.....))..)))))).)))))....))))))))))..))... ( -40.60) >DroYak_CAF1 60463 120 - 1 ACUUUCCGGUUUUCUGGCGUGAACGGCAAAAACCCAGUAAUUUUCAAUAGCAAGGACGUCAGCUGCCGGGCCUGAAAUGUUUAAUAGUGGGCUCGGCGGGUAAAGAGGAUUGGGUAAGGA .(((((((((((((((.((....)).)....((((.............(((..........)))((((((((((..((.....))..))))))))))))))..))))))))))).))).. ( -43.10) >consensus ACUUUCCGGUUUUCUGGCGUGAACGGCAAAAACCCAGUAAUUUUCAAUAGCAAGGACGUCAGCUGCCGGGCCUGAAAUGUUUAAUAGUGGGCUCGGCGGGUAAAGAGGAUUGGGUAAGGA .(((((((((((((((.((....)).)....((((.............(((..........)))((((((((((..((.....))..))))))))))))))..))))))))))).))).. (-39.12 = -39.80 + 0.68)



| Location | 22,066,177 – 22,066,284 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.69 |
| Mean single sequence MFE | -47.24 |
| Consensus MFE | -36.56 |
| Energy contribution | -37.04 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22066177 107 - 27905053 ------------GGCUCUGGUGGAGUGGGUGAAGUGGGUGGAG-CGGGUGGCUCGGUGGCUGGCGGUUAACCCUAAGUGUUGUCACACCACUGCACUCCACACCCUGCUCCAUCACUGCC ------------(((..(((((((((((((...(((((((.((-.(((((((......(((((.((....)))).)))...))))).)).)).))).)))))))).)))))))))..))) ( -51.00) >DroSec_CAF1 58162 98 - 1 ------------GGCUCUGGUGGAGUG---------GGCCGAG-CGGGUGGCUCGGUGGCUGGCGGUUAACCCUAAGUGUUGUCACACCACUGCACUCCACACCCUGCUCCAUCACUGCC ------------(((..((((((((..---------((.((((-(.....)))))((((.(((.((....)))))(((((.((......)).)))))))))..))..))))))))..))) ( -44.80) >DroSim_CAF1 51181 98 - 1 ------------GGCUCUGGUGGAGUG---------GGUGGAC-CGGGUGGCUCGGUGGCUGGCGGUUAACCCUAAGUGUUGUCACACCACUGCACUCCACACCCUGCUCCAUCACUGCC ------------(((..((((((((((---------((((...-.(((((...((((((.(((.((....))))).(((....))).)))))))))))..))))).)))))))))..))) ( -46.60) >DroEre_CAF1 52714 102 - 1 --------UGGUGGCUCUGGUGGAGUG---------GGUGGUU-CGGGUGGCUCGGCGGCAGGCGGUUAACCCUAAGUGUUGUCACACCACUGCACUCCACACCCUGCUCCAUCACUGCC --------....(((..((((((((((---------((((...-.(((((((.((.(....).))))).))))..(((((.((......)).)))))...))))).)))))))))..))) ( -44.60) >DroYak_CAF1 60583 111 - 1 GGUGGGCAUGGUGGCUCUGAUGGAGUG---------GGUGGUGCCGGGUGGCUCAGUGGCUGGCGGUUAACCCUAAGUGUUGUCACACCACUGCACUCCACACCCUGCUCCAUCACUGCC ((((((((.((((.......(((((((---------(((((((((....)))...(((((.((((.(((....))).))))))))))))))).))))))))))).))).)))))...... ( -49.21) >consensus ____________GGCUCUGGUGGAGUG_________GGUGGAG_CGGGUGGCUCGGUGGCUGGCGGUUAACCCUAAGUGUUGUCACACCACUGCACUCCACACCCUGCUCCAUCACUGCC ............(((..((((((((((.........((((.....(((((...((((((.(((.((....))))).(((....))).)))))))))))..)))).))))))))))..))) (-36.56 = -37.04 + 0.48)

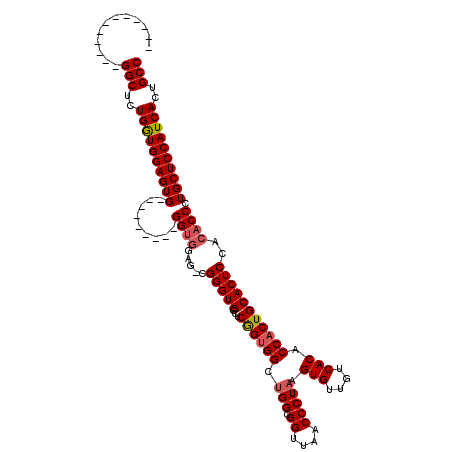
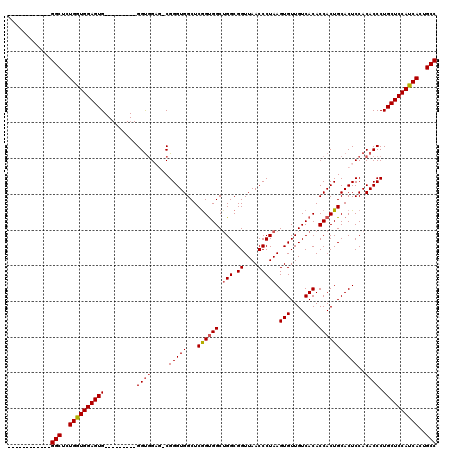
| Location | 22,066,284 – 22,066,389 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.01 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -33.60 |
| Energy contribution | -33.24 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22066284 105 - 27905053 AGCAGGCAGGCAAAAACGCCAUGAACCAUAAAGCAAAGGCAAAGAACAUUCUGGGCGAGAGUGCUCUGGGCUGGGUCGGAGUGGGUGGGACCUGGUGGCUAUGGU--------------- (((.(.(((((..(..(((..(((.((..........(((..(((.((((((.....)))))).)))..))).)))))..)))..)..).)))).).))).....--------------- ( -31.00) >DroSec_CAF1 58260 104 - 1 AGCAGGCAGGCAAAAACGCCAUGAACCAUAAAGCAAAGGCAAAGAACAUUCUGGGCGAGAGUGUUUUGGGCUGGGCCGGAGUGGGUG-GACCCGGUGGCUAUGGU--------------- ........(((......)))....(((((..(((...(((..((((((((((.....))))))))))..)))..(((((.((.....-.))))))).))))))))--------------- ( -35.80) >DroSim_CAF1 51279 104 - 1 AGCAGGCAGGCAAAAACGCCAUGAACCAUAAAGCAAAGGCAAAGAACAUUCUGGGCGAGAGUGUUUUGGGCUGGGCCGGAGUGGGUG-GACCCGGUGGCUAUGGU--------------- ........(((......)))....(((((..(((...(((..((((((((((.....))))))))))..)))..(((((.((.....-.))))))).))))))))--------------- ( -35.80) >DroEre_CAF1 52816 109 - 1 GGCGGGCUGGCAAAAACGCCAUGAACCAUAAAGCAAAGGCAAAGAACAUUCUGGGCGAGAGUGUUUUGGGCUGGGCCGAAGUGGGUG-GACCCGUUGGCUAUGGUGGCAA---------- .....((((((......)))....(((((........(((..((((((((((.....))))))))))..))).((((((...(((..-..))).))))))))))))))..---------- ( -39.90) >DroYak_CAF1 60694 119 - 1 GGCAGGCAGGCAAAAACGCCAUGAACCAUCAAGCAAAGGCAAAGAACAUUCUGGGCGAGAGUGUUUUGGGCUGGGCCGGAAUGGGUG-GACCUGGUGGCUAUGGUGGCCAUGGUGGCUGU .((((.(.(((......))).....(((((..((...(((..((((((((((.....))))))))))..)))..))..).))))...-.(((((((.((....)).)))).)))).)))) ( -45.00) >consensus AGCAGGCAGGCAAAAACGCCAUGAACCAUAAAGCAAAGGCAAAGAACAUUCUGGGCGAGAGUGUUUUGGGCUGGGCCGGAGUGGGUG_GACCCGGUGGCUAUGGU_______________ ........(((......)))....(((((........(((..((((((((((.....))))))))))..))).(((((...((((.....)))).))))))))))............... (-33.60 = -33.24 + -0.36)



| Location | 22,066,309 – 22,066,420 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 97.60 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -31.24 |
| Energy contribution | -31.13 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.31 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.548363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22066309 111 - 27905053 UAGUAGUGGCAGGAUGGAAAACUGGCAUGCAAGCAGGCAGGCAAAAACGCCAUGAACCAUAAAGCAAAGGCAAAGAACAUUCUGGGCGAGAGUGCUCUGGGCUGGGUCGGA ..(((.((.(((.........))).))))).....(((.(((......))).................(((..(((.((((((.....)))))).)))..)))..)))... ( -27.90) >DroSec_CAF1 58284 111 - 1 UAGUAGUGGCAGGAUGGAGAACUGGCAUGCAAGCAGGCAGGCAAAAACGCCAUGAACCAUAAAGCAAAGGCAAAGAACAUUCUGGGCGAGAGUGUUUUGGGCUGGGCCGGA ..(((.((.(((.........))).))))).....(((.(((......))).................(((..((((((((((.....))))))))))..)))..)))... ( -32.20) >DroSim_CAF1 51303 111 - 1 UAGUAGUGGCAGGAUGGAGAACUGGCAUGCAAGCAGGCAGGCAAAAACGCCAUGAACCAUAAAGCAAAGGCAAAGAACAUUCUGGGCGAGAGUGUUUUGGGCUGGGCCGGA ..(((.((.(((.........))).))))).....(((.(((......))).................(((..((((((((((.....))))))))))..)))..)))... ( -32.20) >consensus UAGUAGUGGCAGGAUGGAGAACUGGCAUGCAAGCAGGCAGGCAAAAACGCCAUGAACCAUAAAGCAAAGGCAAAGAACAUUCUGGGCGAGAGUGUUUUGGGCUGGGCCGGA ..(((.((.(((.........))).))))).....(((.(((......))).................(((..((((((((((.....))))))))))..)))..)))... (-31.24 = -31.13 + -0.11)


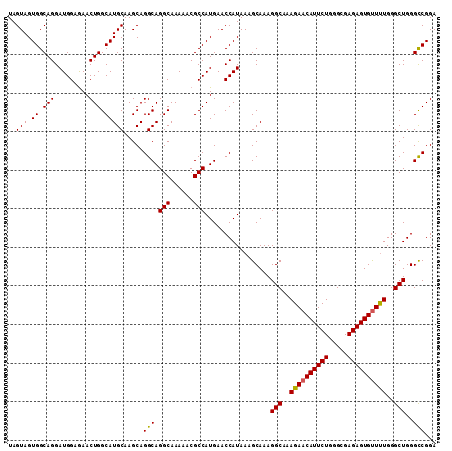
| Location | 22,066,389 – 22,066,495 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 97.48 |
| Mean single sequence MFE | -35.73 |
| Consensus MFE | -33.91 |
| Energy contribution | -34.57 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22066389 106 - 27905053 CACCAUACCAGGCAGGCUAAAAUUCUGGCAUUCUGGGCAGGAUGGCAAACAAGCUAGAAGGCUGGCACGAUGGCGUAGUAGUGGCAGGAUGGAAAACUGGCAUGCA ...(((.((((.....((...((((((.(((((((.((......((.....((((....)))).))......)).))).)))).))))))))....)))).))).. ( -30.80) >DroSec_CAF1 58364 106 - 1 CACGAUGCCAGGCAGGCUAACAUUCUGGCAUUCUGGGCAGGAUGGCAAACAAGCUAGAAGGCUGGCACGAUGGCGUAGUAGUGGCAGGAUGGAGAACUGGCAUGCA ....(((((((.....((..(((((((.(((((((.((......((.....((((....)))).))......)).))).)))).))))))).))..)))))))... ( -38.20) >DroSim_CAF1 51383 106 - 1 CACGAUGCCAGGCAGGCUAACAUUCUGGCAUUCUGGGCAGGAUGGCAAACAAGCUAGAAGGCUGGCACGAUGGCGUAGUAGUGGCAGGAUGGAGAACUGGCAUGCA ....(((((((.....((..(((((((.(((((((.((......((.....((((....)))).))......)).))).)))).))))))).))..)))))))... ( -38.20) >consensus CACGAUGCCAGGCAGGCUAACAUUCUGGCAUUCUGGGCAGGAUGGCAAACAAGCUAGAAGGCUGGCACGAUGGCGUAGUAGUGGCAGGAUGGAGAACUGGCAUGCA ....(((((((.........(((((((.(((((((.((......((.....((((....)))).))......)).))).)))).))))))).....)))))))... (-33.91 = -34.57 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:28 2006