
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,065,714 – 22,065,907 |
| Length | 193 |
| Max. P | 0.986143 |

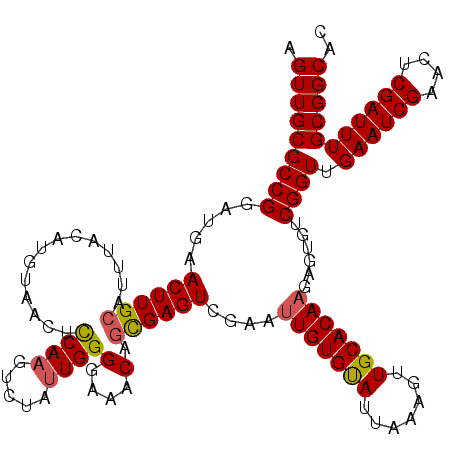
| Location | 22,065,714 – 22,065,834 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -38.78 |
| Consensus MFE | -32.12 |
| Energy contribution | -32.24 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22065714 120 + 27905053 AGUUGCGCCGGAUGAACUUGCAUUUACAUGUAACUCCAAGUCUAUUGGGGAAACAGUGAGUCGAAUUGUGCAUUAAAGUUGCACACGAGUGUCGGUUGAAUCGAACUCGAUUUGCGGCAC .((((((((((....(((((.((((((.(((..((((((.....))))))..))))))))).....((((((.......))))))))))).))))).((((((....))))))))))).. ( -41.80) >DroSec_CAF1 57703 120 + 1 AGUUGCGCCGGAUGAACUUGCAUUUACAUGUAACUCCAAGUCUAUUGGGAAAACAGCGAGUCGAAUUGUGUAUUAAAGUUGCACAAGAGUGUCGGUUGAAUCGAACUCGAUUUGCGGCAC .(((...((.((((.(((((...(((....)))...))))).)))).))..))).(((((((...(((((((.......)))))))((((.(((((...))))))))))))))))..... ( -36.50) >DroSim_CAF1 50712 120 + 1 UGUUGCGCCGGAUGAACUUGCAUUUACAUGUAACUCCAAGUCUAUUGGGGAAACAGCGAGUCGAAUUGUGUAUUAAAGUUGCACAAGAGUGUCGGUUGAAUCGAACUCGAUUUGCGGCAC ......((((((((......)))))...(((..((((((.....))))))..)))(((((((...(((((((.......)))))))((((.(((((...))))))))))))))))))).. ( -38.90) >DroEre_CAF1 52251 120 + 1 AGUUGCGCCGGAUGCACUUGCAUUUGCAUGUAACUUCAAGUCCAUUGGGGAAACAGCGAGUUGAAUUGUGUAUUAAAGUUGCACAAGAGUGGCGGUUGAAUCGAACUCGAUUUGCGGCAC .((((((((.(((((((...(((((((.(((..((((((.....))))))..)))))))).))....)))))))...((..(......)..))))).((((((....))))))))))).. ( -38.70) >DroYak_CAF1 60123 120 + 1 AGUUGCGCCGGAUGAACUUGCAUUUACAUGCAACUCCAAGUCUAGUGGGGAAACAUCGAGUGGAAUUGUGUAUUAAAGUUGCACAAGAAUGGCGGUUGAAUCGAAAUCGAUUUGCGGCAC .(((((((((((((...((((((....))))))(((((.......)))))...))))........(((((((.......)))))))...))))....((((((....))))))))))).. ( -38.00) >consensus AGUUGCGCCGGAUGAACUUGCAUUUACAUGUAACUCCAAGUCUAUUGGGGAAACAGCGAGUCGAAUUGUGUAUUAAAGUUGCACAAGAGUGUCGGUUGAAUCGAACUCGAUUUGCGGCAC .(((((((((.....((((((..............((((.....))))(....).))))))....(((((((.......)))))))......)))).((((((....))))))))))).. (-32.12 = -32.24 + 0.12)


| Location | 22,065,754 – 22,065,874 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.32 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -24.90 |
| Energy contribution | -26.86 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22065754 120 + 27905053 UCUAUUGGGGAAACAGUGAGUCGAAUUGUGCAUUAAAGUUGCACACGAGUGUCGGUUGAAUCGAACUCGAUUUGCGGCACCCCGAAGUGCAGAGAGAAGUCCCUGGAAGAAAGAAGCUAA (((.(..((((....(..((((....((((((.......)))))).((((.(((((...)))))))))))))..).((((......)))).........))))..).))).......... ( -38.70) >DroSec_CAF1 57743 110 + 1 UCUAUUGGGAAAACAGCGAGUCGAAUUGUGUAUUAAAGUUGCACAAGAGUGUCGGUUGAAUCGAACUCGAUUUGCGGCACCCCAAAGUGCAGAGAUAA----------GGAAGAAGCUAC ....(((((......(((((((...(((((((.......)))))))((((.(((((...)))))))))))))))).....))))).............----------............ ( -29.90) >DroSim_CAF1 50752 120 + 1 UCUAUUGGGGAAACAGCGAGUCGAAUUGUGUAUUAAAGUUGCACAAGAGUGUCGGUUGAAUCGAACUCGAUUUGCGGCACCCCAAAGUGCAGAGAUAAGUCCCUGAAAGGAAUAAACUAA (((.(..((((....(((((((...(((((((.......)))))))((((.(((((...)))))))))))))))).((((......)))).........))))..).))).......... ( -39.20) >DroEre_CAF1 52291 120 + 1 UCCAUUGGGGAAACAGCGAGUUGAAUUGUGUAUUAAAGUUGCACAAGAGUGGCGGUUGAAUCGAACUCGAUUUGCGGCACACAGAAGUGCAAAAAGAAGUCCCUGGAAGAAAAAAGCUAA ((((...((((....(((((((...(((((((.......)))))))((((..((((...)))).))))))))))).((((......)))).........))))))))............. ( -35.10) >DroYak_CAF1 60163 119 + 1 UCUAGUGGGGAAACAUCGAGUGGAAUUGUGUAUUAAAGUUGCACAAGAAUGGCGGUUGAAUCGAAAUCGAUUUGCGGCACCCAGGAGUGCAAAAAUAAAUC-CUGGAUAAAAAAAGCUAA ((((.((((....).)))..)))).(((((((.......)))))))...((((.((((.((((....))))...))))..((((((.............))-)))).........)))). ( -30.62) >consensus UCUAUUGGGGAAACAGCGAGUCGAAUUGUGUAUUAAAGUUGCACAAGAGUGUCGGUUGAAUCGAACUCGAUUUGCGGCACCCCGAAGUGCAGAGAUAAGUCCCUGGAAGAAAAAAGCUAA ......(((((....(((((((...(((((((.......)))))))((((.(((((...)))))))))))))))).((((......)))).........)))))................ (-24.90 = -26.86 + 1.96)



| Location | 22,065,794 – 22,065,907 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.57 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -15.18 |
| Energy contribution | -15.78 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22065794 113 + 27905053 GCACACGAGUGUCGGUUGAAUCGAACUCGAUUUGCGGCACCCCGAAGUGCAGAGAGAAGUCCCUGGAAGAAAGAAGCUAAAUUCUAUCCAAAG------AUAGAUAGUAAUG-UAUUGAC ((((.((.((((((...((((((....)))))).))))))..))..)))).........((....))........((((...((((((....)------)))))))))....-....... ( -26.80) >DroSec_CAF1 57783 109 + 1 GCACAAGAGUGUCGGUUGAAUCGAACUCGAUUUGCGGCACCCCAAAGUGCAGAGAUAA----------GGAAGAAGCUACAUUCUAUCUAAAGAGUUAGAUGGAGAAUAAAA-UAUUGCC ((((..(.((((((...((((((....)))))).)))))).)....))))........----------.............((((((((((....)))))))))).......-....... ( -27.20) >DroSim_CAF1 50792 119 + 1 GCACAAGAGUGUCGGUUGAAUCGAACUCGAUUUGCGGCACCCCAAAGUGCAGAGAUAAGUCCCUGAAAGGAAUAAACUAAAUUCUAUACAAAGAGUUAGAUAGAGAAUAAAA-UAUUGAC ((((..(.((((((...((((((....)))))).)))))).)....))))...((((..(((......))).....((((.((((......)))))))).............-))))... ( -23.70) >DroEre_CAF1 52331 117 + 1 GCACAAGAGUGGCGGUUGAAUCGAACUCGAUUUGCGGCACACAGAAGUGCAAAAAGAAGUCCCUGGAAGAAAAAAGCUAAAUUCCAAUCAAAGGGAUAAAUA-GUGAUUAAAAU--GGAG ((((....(((((.((.((((((....)))))))).)).)))....))))........((((((((((.............))))......)))))).....-...........--.... ( -27.72) >DroYak_CAF1 60203 112 + 1 GCACAAGAAUGGCGGUUGAAUCGAAAUCGAUUUGCGGCACCCAGGAGUGCAAAAAUAAAUC-CUGGAUAAAAAAAGCUAAAUUCCAUCCAAAGGAAUAAAUAAGUGAUAAAA-U------ .(((.....((((.((((.((((....))))...))))..((((((.............))-)))).........)))).(((((.......)))))......)))......-.------ ( -26.22) >consensus GCACAAGAGUGUCGGUUGAAUCGAACUCGAUUUGCGGCACCCCGAAGUGCAGAGAUAAGUCCCUGGAAGAAAAAAGCUAAAUUCUAUCCAAAGAGAUAAAUAGAUAAUAAAA_UAUUGAC ((((..(.((((((...((((((....)))))).)))))).)....))))...................................................................... (-15.18 = -15.78 + 0.60)



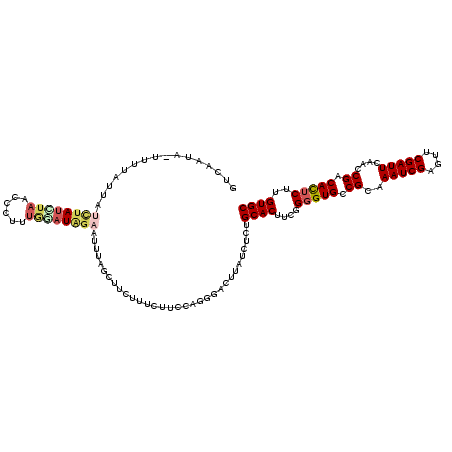
| Location | 22,065,794 – 22,065,907 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.57 |
| Mean single sequence MFE | -24.30 |
| Consensus MFE | -14.50 |
| Energy contribution | -15.78 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22065794 113 - 27905053 GUCAAUA-CAUUACUAUCUAU------CUUUGGAUAGAAUUUAGCUUCUUUCUUCCAGGGACUUCUCUCUGCACUUCGGGGUGCCGCAAAUCGAGUUCGAUUCAACCGACACUCGUGUGC .......-............(------(((((((.((((..........)))))))))))).........(((((....))))).(((...((((((((.......))).)))))..))) ( -27.60) >DroSec_CAF1 57783 109 - 1 GGCAAUA-UUUUAUUCUCCAUCUAACUCUUUAGAUAGAAUGUAGCUUCUUCC----------UUAUCUCUGCACUUUGGGGUGCCGCAAAUCGAGUUCGAUUCAACCGACACUCUUGUGC (((.(((-(((((.......(((((....))))))))))))).)))......----------........((((...((((((.((..(((((....)))))....)).)))))).)))) ( -24.61) >DroSim_CAF1 50792 119 - 1 GUCAAUA-UUUUAUUCUCUAUCUAACUCUUUGUAUAGAAUUUAGUUUAUUCCUUUCAGGGACUUAUCUCUGCACUUUGGGGUGCCGCAAAUCGAGUUCGAUUCAACCGACACUCUUGUGC (((((((-..(((...(((((.(((....))).)))))...)))..))))((.....)))))........((((...((((((.((..(((((....)))))....)).)))))).)))) ( -24.50) >DroEre_CAF1 52331 117 - 1 CUCC--AUUUUAAUCAC-UAUUUAUCCCUUUGAUUGGAAUUUAGCUUUUUUCUUCCAGGGACUUCUUUUUGCACUUCUGUGUGCCGCAAAUCGAGUUCGAUUCAACCGCCACUCUUGUGC ....--...........-......(((((..((..((((........))))..)).))))).........((((....(((.((.(..(((((....)))))...).)))))....)))) ( -21.50) >DroYak_CAF1 60203 112 - 1 ------A-UUUUAUCACUUAUUUAUUCCUUUGGAUGGAAUUUAGCUUUUUUUAUCCAG-GAUUUAUUUUUGCACUCCUGGGUGCCGCAAAUCGAUUUCGAUUCAACCGCCAUUCUUGUGC ------.-.................((((..((((((((.........))))))))))-)).........((((.....((((.....(((((....)))))....))))......)))) ( -23.30) >consensus GUCAAUA_UUUUAUUAUCUAUCUAACCCUUUGGAUAGAAUUUAGCUUCUUUCUUCCAGGGACUUAUCUCUGCACUUCGGGGUGCCGCAAAUCGAGUUCGAUUCAACCGACACUCUUGUGC ................((((((((......))))))))................................((((....(((((.((..(((((....)))))....)).)))))..)))) (-14.50 = -15.78 + 1.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:22 2006