
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,063,898 – 22,064,014 |
| Length | 116 |
| Max. P | 0.949854 |

| Location | 22,063,898 – 22,064,014 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.71 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -26.29 |
| Energy contribution | -26.89 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22063898 116 + 27905053 UUUAUUCGUGAGUGCCAAGGCUAAAGGGGAAAUAUA----UAUAUUUUUGCGUUUCCCAGAAACUGUCAAGUGUGGAUUCGUGUUCAAGCAGGAGCGUCCAACUGACACUCACUAAAUAU .......((((((((....)).....((((((..((----........))..)))))).......((((.((.(((((((.(((....))).))..)))))))))))))))))....... ( -33.20) >DroSec_CAF1 55871 120 + 1 UUUAUUCGUGAGUGCCAAGGCUACAGCGGAAAUAUAUAUAUAUAUUAUUGCGUUUCCCAGAAACUGUCAAGUGUGGAUUCGUGUUCAAGCAGGAGCAUCCAGCUGACACUCACUAAAUAU .......(((((((.(..(((....((((.((((((....)))))).))))(((((...))))).))).(((.(((((((.(((....))).))..))))))))).)))))))....... ( -31.90) >DroSim_CAF1 48882 116 + 1 UUUAUUCGUGAGUGCCAAGGCUAAAGGGGAAAUCUA----UAUAUUAUUGCGUUUCCCAGAAACUGUCAAGUGUGGAUUCGUGUUCAAGCAGGAGCAUCCAGCUGACACUCACUAAAUAU .......((((((((....)).....(((((((.((----........)).))))))).......((((.((.(((((((.(((....))).))..)))))))))))))))))....... ( -32.70) >DroEre_CAF1 50452 115 + 1 UUUAUUCGUAAGUGCCAAGGCUAAAGGGGAAAUAUA----UAGAUUAUU-CGUUUCCCAGAAACUGUCAAGUGUGGAUUCGUGUUCAAGCAGGAGCAUCCAACUGACACUCACUGAAUAU ..(((((((.(((((....)).....(((((((..(----((...))).-.))))))).......((((.((.(((((((.(((....))).))..)))))))))))))).)).))))). ( -29.10) >DroYak_CAF1 58276 116 + 1 UUUAUUCGAAAGUGCCAAGGCUAAAGGGGAAAUAUA----UAGAUUAUUGCGUUUCCCAGAAACUGUCAAGUGUGGAUUCGUGUUCAAGCAGGAGCAUCCAAAUGACACUCACUAAACAU ....(((......((....)).....((((((..((----........))..)))))).)))..(((((....(((((((.(((....))).))..)))))..)))))............ ( -25.70) >consensus UUUAUUCGUGAGUGCCAAGGCUAAAGGGGAAAUAUA____UAUAUUAUUGCGUUUCCCAGAAACUGUCAAGUGUGGAUUCGUGUUCAAGCAGGAGCAUCCAACUGACACUCACUAAAUAU .......((((((((....)).....(((((((..................))))))).......((((.((.(((((((.(((....))).))..)))))))))))))))))....... (-26.29 = -26.89 + 0.60)

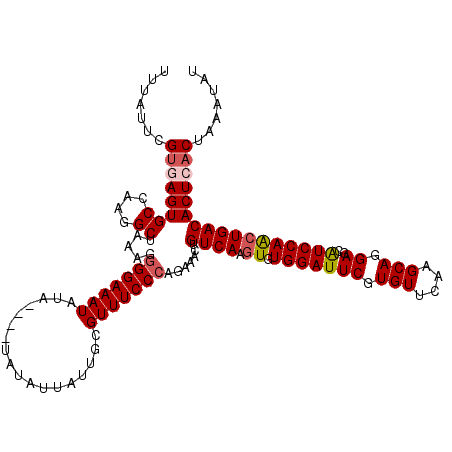
| Location | 22,063,898 – 22,064,014 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.71 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -25.13 |
| Energy contribution | -25.17 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22063898 116 - 27905053 AUAUUUAGUGAGUGUCAGUUGGACGCUCCUGCUUGAACACGAAUCCACACUUGACAGUUUCUGGGAAACGCAAAAAUAUA----UAUAUUUCCCCUUUAGCCUUGGCACUCACGAAUAAA .(((((.(((((((((((((.((.((....)))).)))..................(((...((((((............----....))))))....)))..))))))))))))))).. ( -27.99) >DroSec_CAF1 55871 120 - 1 AUAUUUAGUGAGUGUCAGCUGGAUGCUCCUGCUUGAACACGAAUCCACACUUGACAGUUUCUGGGAAACGCAAUAAUAUAUAUAUAUAUUUCCGCUGUAGCCUUGGCACUCACGAAUAAA .(((((.(((((((((((((((((((....))(((....))))))).......(((((...(((....).))..((((((....))))))...))))))))..))))))))))))))).. ( -30.80) >DroSim_CAF1 48882 116 - 1 AUAUUUAGUGAGUGUCAGCUGGAUGCUCCUGCUUGAACACGAAUCCACACUUGACAGUUUCUGGGAAACGCAAUAAUAUA----UAGAUUUCCCCUUUAGCCUUGGCACUCACGAAUAAA .(((((.(((((((((((((((((((....))(((....)))))))................(((((((...........----..).))))))....)))..))))))))))))))).. ( -29.42) >DroEre_CAF1 50452 115 - 1 AUAUUCAGUGAGUGUCAGUUGGAUGCUCCUGCUUGAACACGAAUCCACACUUGACAGUUUCUGGGAAACG-AAUAAUCUA----UAUAUUUCCCCUUUAGCCUUGGCACUUACGAAUAAA .(((((.(((((((((((((((((((....))(((....)))))))).))......(((...((((((..-.........----....))))))....)))..))))))))))))))).. ( -28.26) >DroYak_CAF1 58276 116 - 1 AUGUUUAGUGAGUGUCAUUUGGAUGCUCCUGCUUGAACACGAAUCCACACUUGACAGUUUCUGGGAAACGCAAUAAUCUA----UAUAUUUCCCCUUUAGCCUUGGCACUUUCGAAUAAA .(((((((((((((((.....)))))))..)).))))))...........((((.(((....((((((............----....)))))).....((....))))).))))..... ( -22.29) >consensus AUAUUUAGUGAGUGUCAGUUGGAUGCUCCUGCUUGAACACGAAUCCACACUUGACAGUUUCUGGGAAACGCAAUAAUAUA____UAUAUUUCCCCUUUAGCCUUGGCACUCACGAAUAAA .(((((.((((((((((..(((((((....))(((....)))))))).........(((...((((((....................))))))....)))..))))))))))))))).. (-25.13 = -25.17 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:13 2006