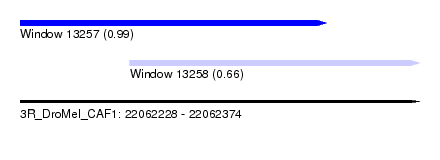
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,062,228 – 22,062,374 |
| Length | 146 |
| Max. P | 0.991215 |

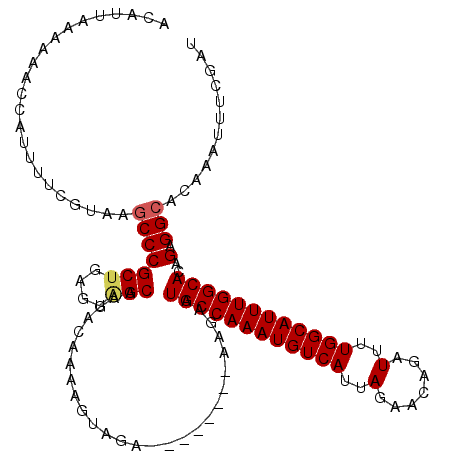
| Location | 22,062,228 – 22,062,340 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.49 |
| Mean single sequence MFE | -25.86 |
| Consensus MFE | -22.80 |
| Energy contribution | -23.08 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22062228 112 + 27905053 ACAUUAAAAAACCAUUUUCGUAUACCCGCUGAGUAGCAAAACAAAAGUAGA--------AAGAAUGCCAAAUGUCAUUAGAACAGAUUUUGGCAUUUGGCACAGAGGCACAAAUUUCGAU ...........((.(((((........(((....)))...((....)).))--------)))..((((((((((((..(.......)..))))))))))))....))............. ( -21.60) >DroSec_CAF1 54214 112 + 1 ACAUUAAAAAACCAUUUUCGUAAGCCCGCUGAGCAGCAAGACAAAAGUAGA--------AAGAAUGCCAAAUGUCAUUAGAACAGAUUUUGGCAUUUGGCACAGAGGCACAAAUUUCGAU .......................(((((((....)))..............--------.....((((((((((((..(.......)..))))))))))))..).)))............ ( -25.20) >DroSim_CAF1 46915 112 + 1 ACAUUAAAAAACCAUUUUCGUAAGCCCGCUGAGCAGCAAGACAAAAGUAGA--------AAGAAUGCCAAAUGUCAUUAGAACAGAUUUUGGCAUUUGGCACAGAGGCACAAAUUUCGAU .......................(((((((....)))..............--------.....((((((((((((..(.......)..))))))))))))..).)))............ ( -25.20) >DroEre_CAF1 48778 112 + 1 ACAUUAAAAAACCAUUUUCGUAAGCCCGCUGAGUUGCGAGACAAGAGUAGA--------AAGAAUGCCAAAUGUCAUUAGAACAGAUUUUGGCAUUUGGCACAGAGGCACAAAUUUCGAU .......................(((..(((.(((....))).........--------.....((((((((((((..(.......)..))))))))))))))).)))............ ( -25.50) >DroYak_CAF1 56555 120 + 1 ACAUUAAAAACCCAUUUUUGUAAGCCCGCUGAGUGGCGAGACAAAAGUAGAAAAGUGGAAAGAAUGCCAAAUGUCAUUAGAACAGAUUUUGGCAUUUGGCACAGAGGCACAAAUUUCGAU .............((((((((..((((.....).)))...)))))))).((((..(((......((((((((((((..(.......)..))))))))))))......).))..))))... ( -31.80) >consensus ACAUUAAAAAACCAUUUUCGUAAGCCCGCUGAGUAGCAAGACAAAAGUAGA________AAGAAUGCCAAAUGUCAUUAGAACAGAUUUUGGCAUUUGGCACAGAGGCACAAAUUUCGAU .......................(((((((....)))...........................((((((((((((..(.......)..))))))))))))..).)))............ (-22.80 = -23.08 + 0.28)

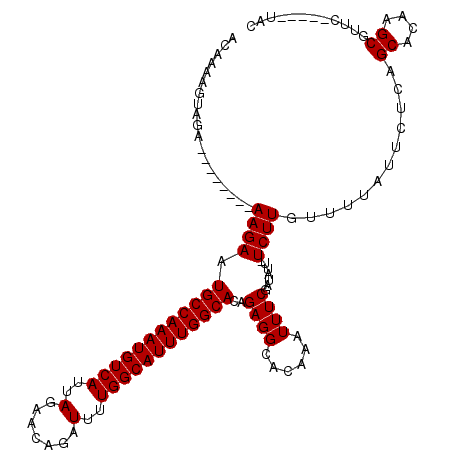
| Location | 22,062,268 – 22,062,374 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.21 |
| Mean single sequence MFE | -26.32 |
| Consensus MFE | -19.44 |
| Energy contribution | -19.44 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22062268 106 + 27905053 ACAAAAGUAGA--------AAGAAUGCCAAAUGUCAUUAGAACAGAUUUUGGCAUUUGGCACAGAGGCACAAAUUUCGAUAUU-UCUUGUUUUAUUCUCAGCACAAGCGUUC-----UAC ......(((((--------(((((((((((((((((..(.......)..)))))))))))...((((((.((((......)))-)..)))))))))))..((....)).)))-----))) ( -26.50) >DroSec_CAF1 54254 106 + 1 ACAAAAGUAGA--------AAGAAUGCCAAAUGUCAUUAGAACAGAUUUUGGCAUUUGGCACAGAGGCACAAAUUUCGAUAUU-UCUUGUUUUAUACUCAGCACAAGCGUUC-----UAC ......(((((--------(....((((((((((((..(.......)..))))))))))))..(((....((((...((....-))..))))....))).((....)).)))-----))) ( -25.80) >DroSim_CAF1 46955 106 + 1 ACAAAAGUAGA--------AAGAAUGCCAAAUGUCAUUAGAACAGAUUUUGGCAUUUGGCACAGAGGCACAAAUUUCGAUAUU-UCUUGUUUUAUUCUCAGCACAAGCGUUC-----UAC ......(((((--------(((((((((((((((((..(.......)..)))))))))))...((((((.((((......)))-)..)))))))))))..((....)).)))-----))) ( -26.50) >DroEre_CAF1 48818 107 + 1 ACAAGAGUAGA--------AAGAAUGCCAAAUGUCAUUAGAACAGAUUUUGGCAUUUGGCACAGAGGCACAAAUUUCGAUAUUUUCUUGUUUUAUUCACAGCACAAGCGGUC-----UAC ....(((((((--------(((((((((((((((((..(.......)..))))))))))))..((((......))))......))))..))))))))...((....))....-----... ( -25.70) >DroYak_CAF1 56595 120 + 1 ACAAAAGUAGAAAAGUGGAAAGAAUGCCAAAUGUCAUUAGAACAGAUUUUGGCAUUUGGCACAGAGGCACAAAUUUCGAUAUUUUCUUGUUUUAUUCUCAGCACAAGCGGUCUGUUCUAC ..............(((((((((.((((((((((((..(.......)..))))))))))))..((((...((((...((.....))..))))...)))).((....))..))).)))))) ( -27.10) >consensus ACAAAAGUAGA________AAGAAUGCCAAAUGUCAUUAGAACAGAUUUUGGCAUUUGGCACAGAGGCACAAAUUUCGAUAUU_UCUUGUUUUAUUCUCAGCACAAGCGUUC_____UAC ...................((((.((((((((((((..(.......)..))))))))))))..((((......)))).......))))............((....))............ (-19.44 = -19.44 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:05 2006