
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,061,267 – 22,061,376 |
| Length | 109 |
| Max. P | 0.914109 |

| Location | 22,061,267 – 22,061,376 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.97 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -21.42 |
| Energy contribution | -20.94 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22061267 109 + 27905053 ACGGGCAAAAUAUCA-AAGAG----------CCAUGGCUUGCCUCAACAACUGACUCACCCCACUGGUCGGGGUAAGUCCAAGUCAAGAAAUGUGAUAAGUACGACUGAUAUUCCAUAUA ...((...(((((((-..(((----------(....)))).........(((((((.(((((.......))))).))))...(((((....).)))).))).....)))))))))..... ( -25.80) >DroSec_CAF1 53305 110 + 1 ACGGGCAAAAUAUCAAAAGAG----------ACAUGGCUUGCCUCAACAACUGACUCACCCCACUGGCCGGGGUAAGUCCAAGUCAAGAAAUUUGAUAAGUACGACUGAUAUUCCAUAUA ...((...(((((((...(((----------.((.....)).)))....(((((((.(((((.......))))).))))...((((((...)))))).))).....)))))))))..... ( -25.50) >DroSim_CAF1 45982 110 + 1 ACGGACAAAAUAUUAAAAGAG----------ACAUGGCUUACCUCAACAACUGACUCACCCCACUGGCCGGGGUAAGUCCAAGUCAAGAAAUGUGAUAAGUGCGACUGAUAUUCCAUAUA ..(((....((((((.....(----------((.(((((((((((..((..((........)).))...)))))))).))).)))......(((.......)))..)))))))))..... ( -23.00) >DroEre_CAF1 47804 109 + 1 ACGGGCAAAAUAUCA-GAGAG----------CCAUGGCUUACCUCAGCAACUGACUCACCCCACCGAUCGGGGUAGGUCCAAGUCAAGAAAUGUGAUAAGUACCACUGAUAUUCCAUAUA ...((...(((((((-(.(((----------(....)))).........((((((..(((((.......)))))..)))...(((((....).)))).)))....))))))))))..... ( -29.40) >DroYak_CAF1 55568 119 + 1 ACGGGCAAAAUAUCA-AAGAGCCAGCCCCAGCCAUGGCUUACCUUAACAACUGACUCACCCCACCGACCGGGGUAAGUCCAAGUCAAGAAAUGUGAUAAGUGCGACUGAUAUUCCAUAUA .((((((...(((((-..((((((((....))..))))))..(((.((....((((.(((((.......))))).))))...)).))).....)))))..)))..)))............ ( -31.10) >consensus ACGGGCAAAAUAUCA_AAGAG__________CCAUGGCUUACCUCAACAACUGACUCACCCCACUGGCCGGGGUAAGUCCAAGUCAAGAAAUGUGAUAAGUACGACUGAUAUUCCAUAUA ..(((....((((((...(................((....))......(((((((.(((((.......))))).))))...((((.......)))).))).)...)))))))))..... (-21.42 = -20.94 + -0.48)

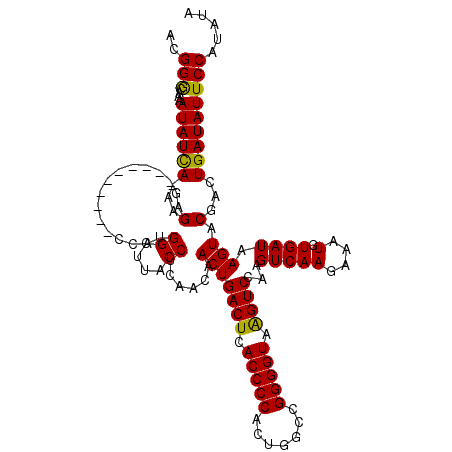
| Location | 22,061,267 – 22,061,376 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.97 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -27.42 |
| Energy contribution | -27.86 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22061267 109 - 27905053 UAUAUGGAAUAUCAGUCGUACUUAUCACAUUUCUUGACUUGGACUUACCCCGACCAGUGGGGUGAGUCAGUUGUUGAGGCAAGCCAUGG----------CUCUU-UGAUAUUUUGCCCGU .....((((((((((................(((..((...(((((((((((.....)))))))))))....))..)))..(((....)----------))..)-)))))))))...... ( -35.30) >DroSec_CAF1 53305 110 - 1 UAUAUGGAAUAUCAGUCGUACUUAUCAAAUUUCUUGACUUGGACUUACCCCGGCCAGUGGGGUGAGUCAGUUGUUGAGGCAAGCCAUGU----------CUCUUUUGAUAUUUUGCCCGU .....((((((((((....................(((...(((((((((((.....)))))))))))....)))((((((.....)))----------)))..))))))))))...... ( -34.10) >DroSim_CAF1 45982 110 - 1 UAUAUGGAAUAUCAGUCGCACUUAUCACAUUUCUUGACUUGGACUUACCCCGGCCAGUGGGGUGAGUCAGUUGUUGAGGUAAGCCAUGU----------CUCUUUUAAUAUUUUGUCCGU .((((((..((((((.....))...........(..((...(((((((((((.....)))))))))))....))..)))))..))))))----------..................... ( -29.60) >DroEre_CAF1 47804 109 - 1 UAUAUGGAAUAUCAGUGGUACUUAUCACAUUUCUUGACUUGGACCUACCCCGAUCGGUGGGGUGAGUCAGUUGCUGAGGUAAGCCAUGG----------CUCUC-UGAUAUUUUGCCCGU .....((((((((((.(((.((((((.((.(..((((((((..((((((......)))))).))))))))..).)).)))))).....)----------))..)-)))))))))...... ( -33.90) >DroYak_CAF1 55568 119 - 1 UAUAUGGAAUAUCAGUCGCACUUAUCACAUUUCUUGACUUGGACUUACCCCGGUCGGUGGGGUGAGUCAGUUGUUAAGGUAAGCCAUGGCUGGGGCUGGCUCUU-UGAUAUUUUGCCCGU .....((((((((((..((............(((((((...(((((((((((.....)))))))))))....)))))))..((((........)))).))...)-)))))))))...... ( -40.60) >consensus UAUAUGGAAUAUCAGUCGUACUUAUCACAUUUCUUGACUUGGACUUACCCCGGCCAGUGGGGUGAGUCAGUUGUUGAGGUAAGCCAUGG__________CUCUU_UGAUAUUUUGCCCGU .....(((((((((...................(((((...(((((((((((.....)))))))))))....)))))((....))....................)))))))))...... (-27.42 = -27.86 + 0.44)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:03 2006