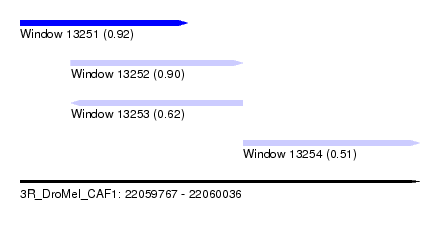
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,059,767 – 22,060,036 |
| Length | 269 |
| Max. P | 0.921474 |

| Location | 22,059,767 – 22,059,880 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.06 |
| Mean single sequence MFE | -36.06 |
| Consensus MFE | -30.32 |
| Energy contribution | -30.52 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22059767 113 + 27905053 CAAUAGGUCAAAGGUCGGAGCCAUCGGCACGGCC-------CAUGCAUAAUGCAUGGAAAAUGUUCGAUUGCCCCUGGCAUUUGUAUCAUUUGCUUUACUAUGCUGCAGGCCAGAAAGCU .....((((...(((((..(((...))).)))))-------..((((....((((((.(((((..(((.((((...)))).)))...)))))......))))))))))))))........ ( -35.60) >DroSec_CAF1 51835 112 + 1 CAAUAGGUCAAAGGUCGGAGCCAUCGGCACGGCC-------CAUGCAUAAUGCAUGGAA-ACGUUCGAUUGCCCCUGGCAUUUGUAUCGUUUGCUUUACUAAGCUGCAGGCCAGAAAGCU .....((((....(((((.((.(((((.(((..(-------(((((.....))))))..-.)))))))).))..))))).........((..((((....)))).)).))))........ ( -36.90) >DroSim_CAF1 44514 113 + 1 CAAUAGGUCAAAGGUCGGAGCCAUCGGCACGGCC-------CAUGCAUAAUGCAUGGAAAAUGUUCGAUUGCCCCUGGCAUUUGUAUCGUUUGCUUUACUAAGCUGCAGGCCAGAAAGCU .....((((...(((((..(((...))).)))))-------(((((.....)))))..(((((..(((.((((...)))).)))...)))))((((....))))....))))........ ( -34.70) >DroEre_CAF1 46298 120 + 1 CAAUAGGUCAAAGGUCGGAGUCAUCGACACGCCCCAUGCCCCAUGCAUAAUGCAUGGAAAAUGUUCGAUUGCCCCUGGCAUUUGUACCAUUUGCUUUAGUAAGCUACCGGCCAGAAAGCU .....(((((((.(((((.((.(((((.((((.....)).((((((.....)))))).....))))))).))..))))).)))).)))....(((((.....((.....))...))))). ( -34.60) >DroYak_CAF1 54134 113 + 1 CAAAAGGCCAAAGGUCGGAGCCAUCGACACGCCC-------CAUGCAUAAUGCAUGGAAAAUGUUCGAUUGCCUCUGGCAUUUGUAUCAUUUGCUUUAGUAAGCUGCAGGCCAGAAAGCU .....((((....((((((((.(((((((....(-------(((((.....))))))....)).))))).).)))))))...((((...(((((....))))).))))))))........ ( -38.50) >consensus CAAUAGGUCAAAGGUCGGAGCCAUCGGCACGGCC_______CAUGCAUAAUGCAUGGAAAAUGUUCGAUUGCCCCUGGCAUUUGUAUCAUUUGCUUUACUAAGCUGCAGGCCAGAAAGCU .....((((...(((((..(((...))).))))).......(((((.....)))))..(((((..(((.((((...)))).)))...)))))((((....))))....))))........ (-30.32 = -30.52 + 0.20)

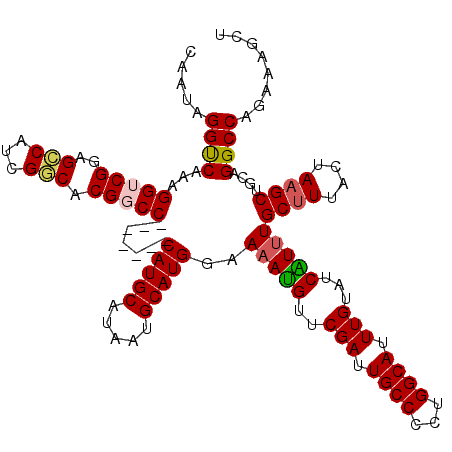
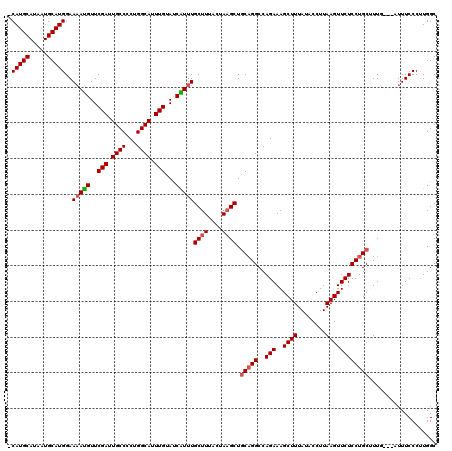
| Location | 22,059,801 – 22,059,917 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.54 |
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -25.50 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22059801 116 + 27905053 -CAUGCAUAAUGCAUGGAAAAUGUUCGAUUGCCCCUGGCAUUUGUAUCAUUUGCUUUACUAUGCUGCAGGCCAGAAAGCUUUAUACCUUAAGUUCUCCUGCUUUG---AUUUCCCUUGGC -(((((.....)))))..(((((..(((.((((...)))).)))...)))))((........)).(((((..(((..((((........))))))))))))....---............ ( -26.60) >DroSec_CAF1 51869 115 + 1 -CAUGCAUAAUGCAUGGAA-ACGUUCGAUUGCCCCUGGCAUUUGUAUCGUUUGCUUUACUAAGCUGCAGGCCAGAAAGCUUUAUACCUUAAGUUCUCCUGCGUUG---AUUUCCCUUGGC -(((((.....))))).((-(((..(((.((((...)))).)))...)))))((((....)))).(((((..(((..((((........))))))))))))((..---(......)..)) ( -31.00) >DroSim_CAF1 44548 116 + 1 -CAUGCAUAAUGCAUGGAAAAUGUUCGAUUGCCCCUGGCAUUUGUAUCGUUUGCUUUACUAAGCUGCAGGCCAGAAAGCUUUAUACCUUAAGUUCUCCUGCGUUG---AUUUCCCUUGGC -(((((.....)))))..(((((..(((.((((...)))).)))...)))))((((....)))).(((((..(((..((((........))))))))))))((..---(......)..)) ( -29.10) >DroEre_CAF1 46338 117 + 1 CCAUGCAUAAUGCAUGGAAAAUGUUCGAUUGCCCCUGGCAUUUGUACCAUUUGCUUUAGUAAGCUACCGGCCAGAAAGCUUUAUACCUUAAGUUCUCCUGCUUUC---AUUUCCCUUGGC ((((((.....)))))).(((((..(((.((((...)))).)))...)))))((((....)))).....((((((((((.....((.....))......))))))---........)))) ( -29.10) >DroYak_CAF1 54168 119 + 1 -CAUGCAUAAUGCAUGGAAAAUGUUCGAUUGCCUCUGGCAUUUGUAUCAUUUGCUUUAGUAAGCUGCAGGCCAGAAAGCUUUAUACCUUAAGUUCUCCUGCUUUCCCCUUUUCCCCUGGC -(((((.....)))))..(((((..(((.((((...)))).)))...)))))((((....)))).(((((..(((..((((........))))))))))))................... ( -28.40) >consensus _CAUGCAUAAUGCAUGGAAAAUGUUCGAUUGCCCCUGGCAUUUGUAUCAUUUGCUUUACUAAGCUGCAGGCCAGAAAGCUUUAUACCUUAAGUUCUCCUGCUUUG___AUUUCCCUUGGC .(((((.....)))))..(((((..(((.((((...)))).)))...)))))((((....)))).(((((..(((..((((........))))))))))))................... (-25.50 = -25.90 + 0.40)

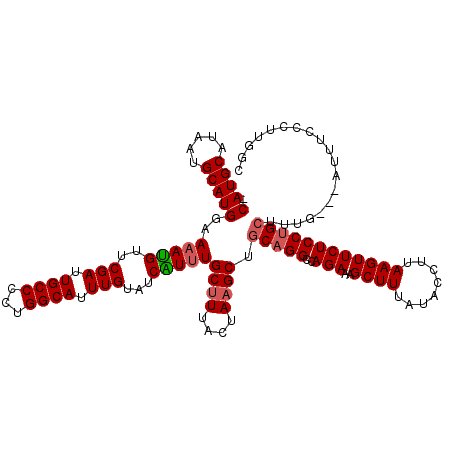
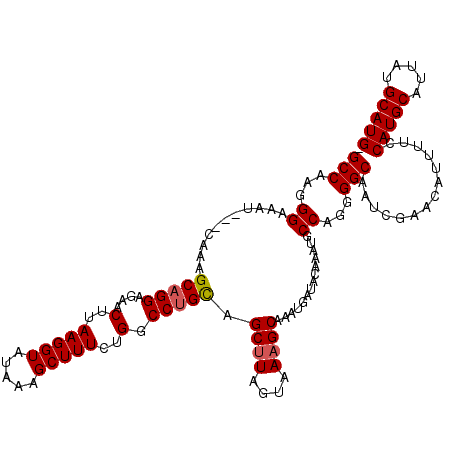
| Location | 22,059,801 – 22,059,917 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.54 |
| Mean single sequence MFE | -28.98 |
| Consensus MFE | -26.74 |
| Energy contribution | -26.98 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22059801 116 - 27905053 GCCAAGGGAAAU---CAAAGCAGGAGAACUUAAGGUAUAAAGCUUUCUGGCCUGCAGCAUAGUAAAGCAAAUGAUACAAAUGCCAGGGGCAAUCGAACAUUUUCCAUGCAUUAUGCAUG- (((...((..((---((..(((((....(..(((((.....)))))..).))))).((........))...)))).......))...)))..............(((((.....)))))- ( -27.20) >DroSec_CAF1 51869 115 - 1 GCCAAGGGAAAU---CAACGCAGGAGAACUUAAGGUAUAAAGCUUUCUGGCCUGCAGCUUAGUAAAGCAAACGAUACAAAUGCCAGGGGCAAUCGAACGU-UUCCAUGCAUUAUGCAUG- ......((((((---....(((((....(..(((((.....)))))..).))))).((((....))))...((((.....((((...))))))))...))-))))((((.....)))).- ( -29.50) >DroSim_CAF1 44548 116 - 1 GCCAAGGGAAAU---CAACGCAGGAGAACUUAAGGUAUAAAGCUUUCUGGCCUGCAGCUUAGUAAAGCAAACGAUACAAAUGCCAGGGGCAAUCGAACAUUUUCCAUGCAUUAUGCAUG- .....(((((((---....(((((....(..(((((.....)))))..).))))).((((....))))...((((.....((((...))))))))...)))))))((((.....)))).- ( -29.80) >DroEre_CAF1 46338 117 - 1 GCCAAGGGAAAU---GAAAGCAGGAGAACUUAAGGUAUAAAGCUUUCUGGCCGGUAGCUUACUAAAGCAAAUGGUACAAAUGCCAGGGGCAAUCGAACAUUUUCCAUGCAUUAUGCAUGG .((..(((((((---(....((((((..(((........))).))))))(((....((((....))))...(((((....)))))..))).......))))))))((((.....)))))) ( -30.50) >DroYak_CAF1 54168 119 - 1 GCCAGGGGAAAAGGGGAAAGCAGGAGAACUUAAGGUAUAAAGCUUUCUGGCCUGCAGCUUACUAAAGCAAAUGAUACAAAUGCCAGAGGCAAUCGAACAUUUUCCAUGCAUUAUGCAUG- (((...((.....(.....(((((....(..(((((.....)))))..).))))).((((....))))........).....))...)))..............(((((.....)))))- ( -27.90) >consensus GCCAAGGGAAAU___CAAAGCAGGAGAACUUAAGGUAUAAAGCUUUCUGGCCUGCAGCUUAGUAAAGCAAAUGAUACAAAUGCCAGGGGCAAUCGAACAUUUUCCAUGCAUUAUGCAUG_ (((...((...........(((((....(..(((((.....)))))..).))))).((((....))))..............))...)))..............(((((.....))))). (-26.74 = -26.98 + 0.24)

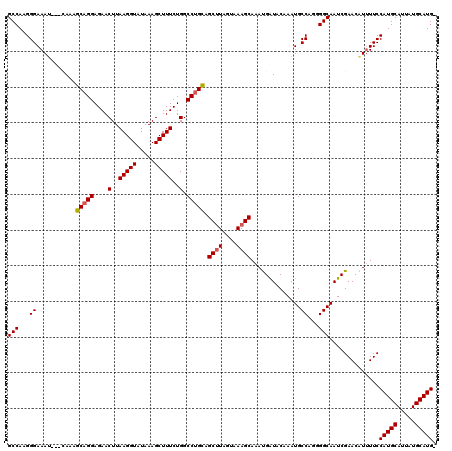
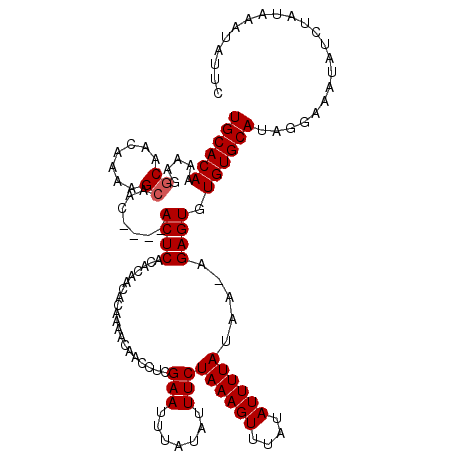
| Location | 22,059,917 – 22,060,036 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.87 |
| Mean single sequence MFE | -15.80 |
| Consensus MFE | -12.10 |
| Energy contribution | -12.30 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.77 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513897 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22059917 119 + 27905053 UGCACAAAAAGGCAACAAAAGCAACACACACUCACACAACACAAAAAAACCUCGAAUUUAUAUUUCUAAAGUUUAUAUUUUAUAA-AGAGUGUGUGCAUAGGAAAUAUCUAUAAAUAUUC (((........)))..........(((((((((....................(((.......)))((((((....))))))...-.))))))))).(((((.....)))))........ ( -18.40) >DroSec_CAF1 51984 115 + 1 UGCACAAAAAGGCAACAAAAGCAAC----ACUCACACAACACAAAACAACCUCGAAUUUAUAUUUCUAAAGUUUAUAUUUUAUAA-AGAGUGUGUGCAUACGAAAUAUCUAUAAAUAUUC (((((.....(....).......((----((((....................(((.......)))((((((....))))))...-.)))))))))))...................... ( -17.50) >DroSim_CAF1 44664 115 + 1 UGCACAAAAAGGCAACAAAAGCAAC----ACUCACACAACACAAAACAACCUCGAAUUUAUAUUUCUAAAGUUUAUAUUUUAUAA-AGAGUGUGUGCAUAGGAAAUAUCUAUAAAUAUUC .((((.....(....).......((----((((....................(((.......)))((((((....))))))...-.))))))))))(((((.....)))))........ ( -17.70) >DroEre_CAF1 46455 114 + 1 UGCACAAA-ACCCAACAAAAGCAAA----ACUCACACAACAAAAAACAACCUCGAAUUUAUAUUUCUAAAGUUUAUAUUUUAUAA-AGAGUGUGUGCAUAGGAAAUAUCUAUAAAUAUUC .(((((..-................----((((....................(((.......)))((((((....))))))...-.)))).)))))(((((.....)))))........ ( -11.31) >DroYak_CAF1 54287 115 + 1 UGCACAGA-ACGCAACAAAAGCAAA----ACUCACACAACAAAAAACAACCUCGAAUUUAUAUUUCUAAAGUUUAUAUUUUAUAAAAGAGUGUGUGCAUAGGAAAUAUCUAUAAAUAUUC .(((((..-..((.......))...----((((....................(((.......)))((((((....)))))).....)))).)))))(((((.....)))))........ ( -14.10) >consensus UGCACAAAAAGGCAACAAAAGCAAC____ACUCACACAACACAAAACAACCUCGAAUUUAUAUUUCUAAAGUUUAUAUUUUAUAA_AGAGUGUGUGCAUAGGAAAUAUCUAUAAAUAUUC ((((((.....((.......)).......((((....................(((.......)))((((((....)))))).....)))).))))))...................... (-12.10 = -12.30 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:06:01 2006