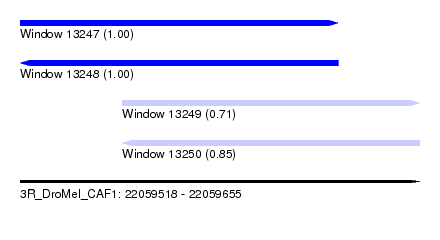
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,059,518 – 22,059,655 |
| Length | 137 |
| Max. P | 0.999862 |

| Location | 22,059,518 – 22,059,627 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -44.84 |
| Consensus MFE | -38.47 |
| Energy contribution | -39.75 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.88 |
| Structure conservation index | 0.86 |
| SVM decision value | 4.29 |
| SVM RNA-class probability | 0.999862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22059518 109 + 27905053 CCGAGCAGUGCUU-UUUCACCUGCCCAACUGGAG----CGUCUGUUGCAGGCGGCAUUUUCAUAUUGAAAAUGCAUUUUGGCCAGUGCACAGUUG---GUUUUCCAGUUGGCCAGAA--- ..((((...))))-......(((.((((((((((----((.((((.((((((.((((((((.....))))))))......)))..))))))).))---...)))))))))).)))..--- ( -39.70) >DroSec_CAF1 51586 109 + 1 CCGAGCAGUGCUU-UUCCACCUGGCCAACUGGAG----CGACUGUUGCAGGCGGCAUUUUCAUAUUGAAAAUGCAUUUUGGCCAGUGCACAGUUC---GUUUUCCAGUUGGCCAUAA--- ..((((...))))-.......(((((((((((((----(((((((.((((((.((((((((.....))))))))......)))..))))))).))---))..))))))))))))...--- ( -48.20) >DroSim_CAF1 44265 109 + 1 CCGAGCAGUGCUU-UUCCACCUGGCCAACUGGAG----CGACUGUUGCAGGCGGCAUUUUCAUAUUGAAAAUGCAUUUUGGCCAGUGCACAGUUG---GCUUUCCAGUUGGCCAGAA--- ..((((...))))-......((((((((((((((----(((((((.((((((.((((((((.....))))))))......)))..))))))))))---...))))))))))))))..--- ( -51.40) >DroEre_CAF1 46041 112 + 1 CCGAGCAGUGCUU-UUCCACUUGGCCAACUGGUG----CGUAUUUCGCAGGCGUCAUUUUCAUAUUGAAAAUGCAUUUUGGCCAGUGCACAGUUG---GUUUUCCAGUUGGCCAGAAUGC ....((((((...-...)))((((((((((((((----((.....))))((((((((((((.....))))))).))....)))............---.....))))))))))))..))) ( -37.90) >DroYak_CAF1 53855 119 + 1 CCGAGCAGUGCUUUUUCCACCUGGCCAACUGGAGCGAGCGUCUUUCGCAGGCGGCAUUUUCAUAUUGAAAAUGCAUUUUGGCCAGUGCACAGUUGGUGGUUUUCCAGUUGGCCAGAAA-C ..((((...)))).......((((((((((((((((((.....))))).(((.((((((((.....))))))))......)))....(((.....)))....)))))))))))))...-. ( -47.00) >consensus CCGAGCAGUGCUU_UUCCACCUGGCCAACUGGAG____CGUCUGUUGCAGGCGGCAUUUUCAUAUUGAAAAUGCAUUUUGGCCAGUGCACAGUUG___GUUUUCCAGUUGGCCAGAA___ ..((((...)))).......((((((((((((((.......((((.((((((.((((((((.....))))))))......)))..))))))).........))))))))))))))..... (-38.47 = -39.75 + 1.28)

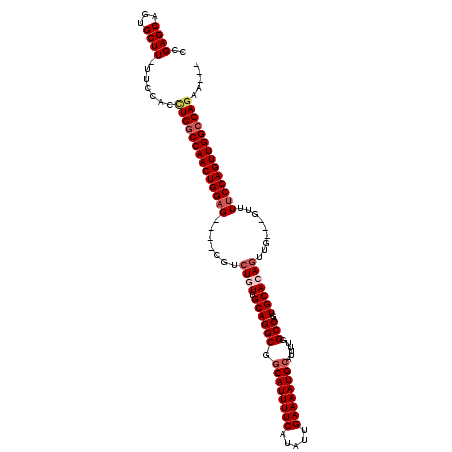
| Location | 22,059,518 – 22,059,627 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -43.38 |
| Consensus MFE | -36.73 |
| Energy contribution | -36.93 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.30 |
| Structure conservation index | 0.85 |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22059518 109 - 27905053 ---UUCUGGCCAACUGGAAAAC---CAACUGUGCACUGGCCAAAAUGCAUUUUCAAUAUGAAAAUGCCGCCUGCAACAGACG----CUCCAGUUGGGCAGGUGAAA-AAGCACUGCUCGG ---..(((.(((((((((....---(..(((((((..(((......((((((((.....)))))))).)))))).))))..)----.))))))))).)))(((...-...)))....... ( -39.70) >DroSec_CAF1 51586 109 - 1 ---UUAUGGCCAACUGGAAAAC---GAACUGUGCACUGGCCAAAAUGCAUUUUCAAUAUGAAAAUGCCGCCUGCAACAGUCG----CUCCAGUUGGCCAGGUGGAA-AAGCACUGCUCGG ---...((((((((((((...(---((.(((((((..(((......((((((((.....)))))))).)))))).)))))))----.))))))))))))((((...-...))))...... ( -47.30) >DroSim_CAF1 44265 109 - 1 ---UUCUGGCCAACUGGAAAGC---CAACUGUGCACUGGCCAAAAUGCAUUUUCAAUAUGAAAAUGCCGCCUGCAACAGUCG----CUCCAGUUGGCCAGGUGGAA-AAGCACUGCUCGG ---..(((((((((((((..((---..((((((((..(((......((((((((.....)))))))).)))))).))))).)----))))))))))))))(((...-...)))....... ( -48.30) >DroEre_CAF1 46041 112 - 1 GCAUUCUGGCCAACUGGAAAAC---CAACUGUGCACUGGCCAAAAUGCAUUUUCAAUAUGAAAAUGACGCCUGCGAAAUACG----CACCAGUUGGCCAAGUGGAA-AAGCACUGCUCGG ((....(((((((((((.....---............(((.......(((((((.....)))))))..)))((((.....))----)))))))))))))((((...-...)))))).... ( -38.90) >DroYak_CAF1 53855 119 - 1 G-UUUCUGGCCAACUGGAAAACCACCAACUGUGCACUGGCCAAAAUGCAUUUUCAAUAUGAAAAUGCCGCCUGCGAAAGACGCUCGCUCCAGUUGGCCAGGUGGAAAAAGCACUGCUCGG .-...(((((((((((((...........((.((....))))....((((((((.....)))))))).((..(((.....)))..)))))))))))))))(..(........)..).... ( -42.70) >consensus ___UUCUGGCCAACUGGAAAAC___CAACUGUGCACUGGCCAAAAUGCAUUUUCAAUAUGAAAAUGCCGCCUGCAACAGACG____CUCCAGUUGGCCAGGUGGAA_AAGCACUGCUCGG ......((((((((((((.............((((..(((......((((((((.....)))))))).)))))))............))))))))))))((((.......))))...... (-36.73 = -36.93 + 0.20)



| Location | 22,059,553 – 22,059,655 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.96 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -24.73 |
| Energy contribution | -25.13 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22059553 102 + 27905053 UCUGUUGCAGGCGGCAUUUUCAUAUUGAAAAUGCAUUUUGGCCAGUGCACAGUUG---GUUUUCCAGUUGGCCAGAA---------------UGUCAGUCAGCAGUUAGUUAGCCUUUGG .(((((((.(((.((((((((.....))))))))(((((((((((.......(((---(....))))))))))))))---------------)))).).))))))............... ( -34.81) >DroSec_CAF1 51621 102 + 1 ACUGUUGCAGGCGGCAUUUUCAUAUUGAAAAUGCAUUUUGGCCAGUGCACAGUUC---GUUUUCCAGUUGGCCAUAA---------------UGUCAGUCAGCAGUUAGUUAGCCUUUGG ((((((((.(((.((((((((.....))))))))(((.(((((((((.((.....---))....)).))))))).))---------------)))).).))))))).............. ( -30.00) >DroSim_CAF1 44300 102 + 1 ACUGUUGCAGGCGGCAUUUUCAUAUUGAAAAUGCAUUUUGGCCAGUGCACAGUUG---GCUUUCCAGUUGGCCAGAA---------------UGUCAGUCAGCAGUUAGUUAGCCUUUGG ((((((((.(((.((((((((.....))))))))(((((((((((((...((...---.))...)).))))))))))---------------)))).).))))))).............. ( -35.30) >DroEre_CAF1 46076 110 + 1 UAUUUCGCAGGCGUCAUUUUCAUAUUGAAAAUGCAUUUUGGCCAGUGCACAGUUG---GUUUUCCAGUUGGCCAGAAUGCCAGAA-------UGUCAGUCAGCAGUUAGUUCGCCUUUGG .....((.(((((((((((((.....))))))(((((((((((((.......(((---(....)))))))))))))))))..)).-------(((......))).......))))).)). ( -32.81) >DroYak_CAF1 53895 119 + 1 UCUUUCGCAGGCGGCAUUUUCAUAUUGAAAAUGCAUUUUGGCCAGUGCACAGUUGGUGGUUUUCCAGUUGGCCAGAAA-CCAGCAGUCCAGCAGUCAGUCAGCAGUUAGUUAGCCUUUGG .....((.((((.((((((((.....))))))))...(((((.(.(((....(((.(((((((((....))...))))-))).)))....))).)..)))))..........)))).)). ( -33.80) >consensus UCUGUUGCAGGCGGCAUUUUCAUAUUGAAAAUGCAUUUUGGCCAGUGCACAGUUG___GUUUUCCAGUUGGCCAGAA_______________UGUCAGUCAGCAGUUAGUUAGCCUUUGG ........((((.((((((((.....)))))))).((((((((((((.................)).))))))))))...................................)))).... (-24.73 = -25.13 + 0.40)



| Location | 22,059,553 – 22,059,655 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.96 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -21.68 |
| Energy contribution | -21.72 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22059553 102 - 27905053 CCAAAGGCUAACUAACUGCUGACUGACA---------------UUCUGGCCAACUGGAAAAC---CAACUGUGCACUGGCCAAAAUGCAUUUUCAAUAUGAAAAUGCCGCCUGCAACAGA (((..(((((..................---------------...)))))...))).....---...(((((((..(((......((((((((.....)))))))).)))))).)))). ( -24.90) >DroSec_CAF1 51621 102 - 1 CCAAAGGCUAACUAACUGCUGACUGACA---------------UUAUGGCCAACUGGAAAAC---GAACUGUGCACUGGCCAAAAUGCAUUUUCAAUAUGAAAAUGCCGCCUGCAACAGU (((..(((((..(((.(((.....).))---------------))))))))...))).....---..((((((((..(((......((((((((.....)))))))).)))))).))))) ( -25.90) >DroSim_CAF1 44300 102 - 1 CCAAAGGCUAACUAACUGCUGACUGACA---------------UUCUGGCCAACUGGAAAGC---CAACUGUGCACUGGCCAAAAUGCAUUUUCAAUAUGAAAAUGCCGCCUGCAACAGU .....((((.......(((.....).))---------------(((..(....)..))))))---).((((((((..(((......((((((((.....)))))))).)))))).))))) ( -27.30) >DroEre_CAF1 46076 110 - 1 CCAAAGGCGAACUAACUGCUGACUGACA-------UUCUGGCAUUCUGGCCAACUGGAAAAC---CAACUGUGCACUGGCCAAAAUGCAUUUUCAAUAUGAAAAUGACGCCUGCGAAAUA ....(((((.................((-------((.((((......((((..(((....)---))..)).))....)))).))))(((((((.....))))))).)))))........ ( -28.70) >DroYak_CAF1 53895 119 - 1 CCAAAGGCUAACUAACUGCUGACUGACUGCUGGACUGCUGG-UUUCUGGCCAACUGGAAAACCACCAACUGUGCACUGGCCAAAAUGCAUUUUCAAUAUGAAAAUGCCGCCUGCGAAAGA ....((((....................((..(..((((((-((((..(....)..).))))))........))))..))......((((((((.....)))))))).)))).(....). ( -28.90) >consensus CCAAAGGCUAACUAACUGCUGACUGACA_______________UUCUGGCCAACUGGAAAAC___CAACUGUGCACUGGCCAAAAUGCAUUUUCAAUAUGAAAAUGCCGCCUGCAACAGA ....((((......................................((((((....(...((........)).)..))))))....((((((((.....)))))))).))))........ (-21.68 = -21.72 + 0.04)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:57 2006