
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,058,263 – 22,058,780 |
| Length | 517 |
| Max. P | 0.991714 |

| Location | 22,058,263 – 22,058,383 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -47.24 |
| Consensus MFE | -43.36 |
| Energy contribution | -43.93 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.689345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22058263 120 + 27905053 GUUCGCCAUUCAUCUUCGCCUACAGGAACAAGAGGGUGAGGCGAGGAGUCAAGCGGCUGUUUGGUCUGGAUUCGUCGUCGGGCCUGCAGCGCAACUGCAGCAGCAGUGUAAAGACCAAUG .((((((..((((((((.((....)).....))))))))))))))(.(((..(((.((((..(((((((........))))))).))))))).(((((....))))).....)))).... ( -47.70) >DroSec_CAF1 50339 120 + 1 GUUCGCCAUUCAUCUUCGCCUACAGAAACAAGAGGGUGAGGCGAGGAGUCAAGCGGCUGUUUGGUCUGGAUUCGUCGUCGGGCCUGCAGCGCAACUGCAGCAGCAGUGUAAAGACCAAUG .((((((..((((((((..............))))))))))))))(.(((..(((.((((..(((((((........))))))).))))))).(((((....))))).....)))).... ( -45.44) >DroSim_CAF1 43023 120 + 1 GUUCGCCAUUCAUCUUCGCCUACAGGAACAAGAGGGUGAGGCGAGGAGUCAAGCGGCUGUUUGGUCUGGAUUCGUCGUCGGGCCUGCAGCGCAACUGCAGCAGCAGUGUAAAGACCAAUG .((((((..((((((((.((....)).....))))))))))))))(.(((..(((.((((..(((((((........))))))).))))))).(((((....))))).....)))).... ( -47.70) >DroEre_CAF1 44789 120 + 1 GUUCGCCCUUCAUCUUCGCCUACAGGAAUAAGAGGGUGCGGCGAGGAGUCAAGCGGCUGUUUGGUCUGCACUCGUCGUCGGGUCUGCAGCGCAACUGCAGCAGCAGUGUGAAGACCAAUG ...(((((((........((....)).....)))))))((((((.((((...((((((....)).)))))))).))))))(((((....((((.((((....)))))))).))))).... ( -48.12) >consensus GUUCGCCAUUCAUCUUCGCCUACAGGAACAAGAGGGUGAGGCGAGGAGUCAAGCGGCUGUUUGGUCUGGAUUCGUCGUCGGGCCUGCAGCGCAACUGCAGCAGCAGUGUAAAGACCAAUG .((((((..((((((((.((....)).....))))))))))))))(.(((..(((.((((..(((((((........))))))).))))))).(((((....))))).....)))).... (-43.36 = -43.93 + 0.56)



| Location | 22,058,343 – 22,058,463 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -51.17 |
| Consensus MFE | -50.91 |
| Energy contribution | -50.35 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22058343 120 + 27905053 GGCCUGCAGCGCAACUGCAGCAGCAGUGUAAAGACCAAUGGCACCGCAGGUCCUGCUGCAAGUGGUGCACAAUUGCAGCGCAACAGCAGCAAGUUGUCGCAGUACAGCAGCAACAGCUGC (..((((...((((((((.(((((((......((((..((....))..)))))))))))..(((.((((....)))).))).......)).)))))).))))..).(((((....))))) ( -49.10) >DroSec_CAF1 50419 120 + 1 GGCCUGCAGCGCAACUGCAGCAGCAGUGUAAAGACCAAUGGCACCACGGGUCCUGCUGCAAGUGGUGCACAAUUGCAGCGCAACAGCAGCAAGUUGUCGCAGUACAGCAGCAACAGCUGC (..((((...((((((((.(((((((......((((..((....))..)))))))))))..(((.((((....)))).))).......)).)))))).))))..).(((((....))))) ( -50.80) >DroSim_CAF1 43103 120 + 1 GGCCUGCAGCGCAACUGCAGCAGCAGUGUAAAGACCAAUGGCACCGCGGGUCCUGCUGCAAGUGGUGCACAAUUGCAGCGCAACAGCAGCAAGUUGUCGCAGUACAGCAGCAACAGCUGC (..((((...((((((((.(((((((......((((..((....))..)))))))))))..(((.((((....)))).))).......)).)))))).))))..).(((((....))))) ( -50.10) >DroEre_CAF1 44869 120 + 1 GGUCUGCAGCGCAACUGCAGCAGCAGUGUGAAGACCAAUGGCACCGCGGGUCCUGCUGCGGGUGGUGCACAGUUGCAGCGCAACAGCAGCAAGCUGUCGCAGUACAGCAGCAACAGCUGC (((((....((((.((((....)))))))).))))).......((((((......))))))(((.(((((((((((.((......)).)).)))))).))).))).(((((....))))) ( -54.70) >consensus GGCCUGCAGCGCAACUGCAGCAGCAGUGUAAAGACCAAUGGCACCGCGGGUCCUGCUGCAAGUGGUGCACAAUUGCAGCGCAACAGCAGCAAGUUGUCGCAGUACAGCAGCAACAGCUGC .....(((((....((((.(((((((......((((..((....))..)))))))))))..(((.(((((((((((.((......)).)).)))))).))).))).)))).....))))) (-50.91 = -50.35 + -0.56)


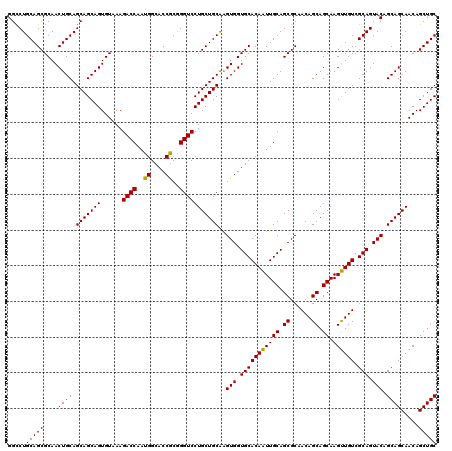
| Location | 22,058,343 – 22,058,463 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -52.85 |
| Consensus MFE | -50.71 |
| Energy contribution | -50.15 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22058343 120 - 27905053 GCAGCUGUUGCUGCUGUACUGCGACAACUUGCUGCUGUUGCGCUGCAAUUGUGCACCACUUGCAGCAGGACCUGCGGUGCCAUUGGUCUUUACACUGCUGCUGCAGUUGCGCUGCAGGCC (((((.((((.(((......))).))))..)))))......(((((((.((.....)).))))))).((.((((((((((.....((....))(((((....))))).)))))))))))) ( -53.70) >DroSec_CAF1 50419 120 - 1 GCAGCUGUUGCUGCUGUACUGCGACAACUUGCUGCUGUUGCGCUGCAAUUGUGCACCACUUGCAGCAGGACCCGUGGUGCCAUUGGUCUUUACACUGCUGCUGCAGUUGCGCUGCAGGCC (((((.((((.(((......))).))))..)))))((.(((((.......))))).))((((((((((((((.(((....))).))))))...(((((....)))))...)))))))).. ( -53.50) >DroSim_CAF1 43103 120 - 1 GCAGCUGUUGCUGCUGUACUGCGACAACUUGCUGCUGUUGCGCUGCAAUUGUGCACCACUUGCAGCAGGACCCGCGGUGCCAUUGGUCUUUACACUGCUGCUGCAGUUGCGCUGCAGGCC (((((....))))).((((((((....((((((((((.(((((.......))))).))...))))))))...))))))))..............((((.((.((....)))).))))... ( -50.90) >DroEre_CAF1 44869 120 - 1 GCAGCUGUUGCUGCUGUACUGCGACAGCUUGCUGCUGUUGCGCUGCAACUGUGCACCACCCGCAGCAGGACCCGCGGUGCCAUUGGUCUUCACACUGCUGCUGCAGUUGCGCUGCAGACC (((((....))))).((((((((....((((((((((.(((((.......))))).))...))))))))...))))))))....(((((.((((((((....)))))...).)).))))) ( -53.30) >consensus GCAGCUGUUGCUGCUGUACUGCGACAACUUGCUGCUGUUGCGCUGCAAUUGUGCACCACUUGCAGCAGGACCCGCGGUGCCAUUGGUCUUUACACUGCUGCUGCAGUUGCGCUGCAGGCC (((((....))))).((((((((....((((((((((.(((((.......))))).))...))))))))...))))))))....((((.....(((((....)))))(((...))))))) (-50.71 = -50.15 + -0.56)



| Location | 22,058,383 – 22,058,500 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 94.02 |
| Mean single sequence MFE | -41.77 |
| Consensus MFE | -37.79 |
| Energy contribution | -37.60 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22058383 117 + 27905053 GCACCGCAGGUCCUGCUGCAAGUGGUGCACAAUUGCAGCGCAACAGCAGCAAGUUGUCGCAGUACAGCAGCAACAGCUGCAAGUAUCUCACCCCCCAGAGUUCCCUGGUCAGUCAAG (((((((..((......))..)))))))....(((((((((....)).....(((((.((......)).))))).)))))))............((((......))))......... ( -39.10) >DroSec_CAF1 50459 117 + 1 GCACCACGGGUCCUGCUGCAAGUGGUGCACAAUUGCAGCGCAACAGCAGCAAGUUGUCGCAGUACAGCAGCAACAGCUGCAAGUACCUCACCCCUCAGAGCUCACUGGUCAGUCAAG (.((((.(((..(((((((((.((.....)).)))))))((.(((((.....))))).)).((((.(((((....)))))..))))..........))..)))..)))))....... ( -39.80) >DroSim_CAF1 43143 117 + 1 GCACCGCGGGUCCUGCUGCAAGUGGUGCACAAUUGCAGCGCAACAGCAGCAAGUUGUCGCAGUACAGCAGCAACAGCUGCAAGUACCUCACCCCCCAGAGCUCACUGGUCAGUCAAG .......((((...(((((((.((.....)).)))))))((.(((((.....))))).)).((((.(((((....)))))..))))...)))).((((......))))......... ( -40.60) >DroEre_CAF1 44909 117 + 1 GCACCGCGGGUCCUGCUGCGGGUGGUGCACAGUUGCAGCGCAACAGCAGCAAGCUGUCGCAGUACAGCAGCAACAGCUGCAGGUACCUCACCCCACAAAGCUCCCUGGUCGGUCAAG (.((((.(((..((..((.(((((((((((((((((.((......)).)).)))))).)))((((.(((((....)))))..)))).))))))))...))..))))))))....... ( -47.60) >consensus GCACCGCGGGUCCUGCUGCAAGUGGUGCACAAUUGCAGCGCAACAGCAGCAAGUUGUCGCAGUACAGCAGCAACAGCUGCAAGUACCUCACCCCCCAGAGCUCACUGGUCAGUCAAG (.((((.(((..(((((((((.((.....)).)))))))((.(((((.....))))).)).((((.(((((....)))))..))))............))..))))))))....... (-37.79 = -37.60 + -0.19)



| Location | 22,058,383 – 22,058,500 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 94.02 |
| Mean single sequence MFE | -50.22 |
| Consensus MFE | -45.88 |
| Energy contribution | -45.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22058383 117 - 27905053 CUUGACUGACCAGGGAACUCUGGGGGGUGAGAUACUUGCAGCUGUUGCUGCUGUACUGCGACAACUUGCUGCUGUUGCGCUGCAAUUGUGCACCACUUGCAGCAGGACCUGCGGUGC (((.(((..(((((....)))))..))))))......(((((....))))).((((((((....((((((((((.(((((.......))))).))...))))))))...)))))))) ( -48.80) >DroSec_CAF1 50459 117 - 1 CUUGACUGACCAGUGAGCUCUGAGGGGUGAGGUACUUGCAGCUGUUGCUGCUGUACUGCGACAACUUGCUGCUGUUGCGCUGCAAUUGUGCACCACUUGCAGCAGGACCCGUGGUGC ..(.(((....))).)((.((..(((((..(((((..(((((....))))).)))))...))..((((((((((.(((((.......))))).))...)))))))).)))..)).)) ( -44.80) >DroSim_CAF1 43143 117 - 1 CUUGACUGACCAGUGAGCUCUGGGGGGUGAGGUACUUGCAGCUGUUGCUGCUGUACUGCGACAACUUGCUGCUGUUGCGCUGCAAUUGUGCACCACUUGCAGCAGGACCCGCGGUGC (((.(((..((((......))))..))).))).....(((((....))))).((((((((....((((((((((.(((((.......))))).))...))))))))...)))))))) ( -51.60) >DroEre_CAF1 44909 117 - 1 CUUGACCGACCAGGGAGCUUUGUGGGGUGAGGUACCUGCAGCUGUUGCUGCUGUACUGCGACAGCUUGCUGCUGUUGCGCUGCAACUGUGCACCACCCGCAGCAGGACCCGCGGUGC ....((((....(((..(((((((((.((..((((..(((((....)))))((((.(((((((((.....))))))))).))))...))))..))))))))).))..))).)))).. ( -55.70) >consensus CUUGACUGACCAGGGAGCUCUGGGGGGUGAGGUACUUGCAGCUGUUGCUGCUGUACUGCGACAACUUGCUGCUGUUGCGCUGCAAUUGUGCACCACUUGCAGCAGGACCCGCGGUGC (((.(((..((((......))))..))).))).....(((((....))))).((((((((....((((((((((.(((((.......))))).))...))))))))...)))))))) (-45.88 = -45.88 + 0.00)

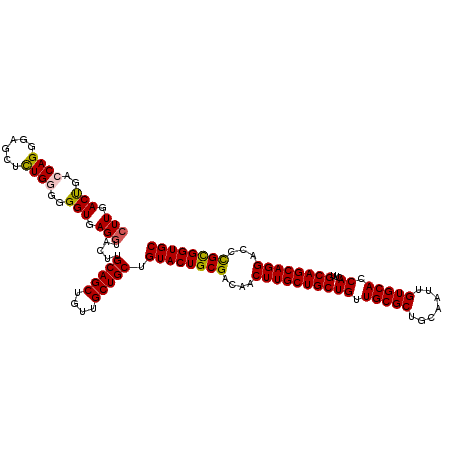

| Location | 22,058,500 – 22,058,620 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -42.30 |
| Consensus MFE | -40.10 |
| Energy contribution | -40.06 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22058500 120 + 27905053 UGCCGGUGCAUACGACCUUGACGUUAAGGCCCAACAGCAGCUGCAGCACCAUCAUCAAUUUCGGUGGCGCCAGGGGCUCAGCGGACUCCGAUGAGCAGCCACCGGCAACACCACCACCUA (((((((((......(((((....)))))(((....((....)).((.(((((.........))))).))..)))(((((.(((...))).))))).).))))))))............. ( -44.20) >DroSec_CAF1 50576 120 + 1 UGCCGGUGCAUACGACCUUGACAUUAAGGCCCAACAGCAGCUGCAGCACCAUCAUCAAUUUCGGUGGCGCCAGGGGCUCAGCGGACUCCGAUGAGCAGCCACCGGCAACACCACCACCCA (((((((((......(((((....)))))(((....((....)).((.(((((.........))))).))..)))(((((.(((...))).))))).).))))))))............. ( -44.20) >DroSim_CAF1 43260 120 + 1 UGCCGGUGCAUACGACCUUGACGUUAAGGCCCAACAGCAGCUGCAGCACCAUCAUCAAUUUCGGUGGCGCCAGGGGCUCAGCGGACUCCGAUGAGCAGCCACCGGCAACACCACCCCCCA (((((((((......(((((....)))))(((....((....)).((.(((((.........))))).))..)))(((((.(((...))).))))).).))))))))............. ( -44.20) >DroEre_CAF1 45026 120 + 1 UGCCGGUGCACACGACCUUGACUUUGAGGCCCAACAGCAGCUGCAGCACCAUCAUCAAUUUCGGUGGCGCCAGGGGUUCAGCGGACUCCGAUGAGCAGCCACCGGCCACACCACCACCCA .((((((((......((((......)))).......((....)).)))))............((((((((((((((((.....))))))..)).)).))))))))).............. ( -42.70) >DroYak_CAF1 52821 120 + 1 UGCCCGUGCACACGACCUUGACCUUGAGGCCCAACAGCAGCUGCAGCACUAUCAUCAAUUUCGGCGGUGCCAGGGGUUCAGCGGACUCCGAUGAGCAGCCACCGGCAACACCACCACCCA ((((.(((.......((((......))))..........(((((.(((((..(.........)..)))))..((((((.....)))))).....)))))))).))))............. ( -36.20) >consensus UGCCGGUGCAUACGACCUUGACGUUAAGGCCCAACAGCAGCUGCAGCACCAUCAUCAAUUUCGGUGGCGCCAGGGGCUCAGCGGACUCCGAUGAGCAGCCACCGGCAACACCACCACCCA (((((((((......(((((....)))))(((....((....)).((.(((((.........))))).))..)))(((((.(((...))).))))).).))))))))............. (-40.10 = -40.06 + -0.04)

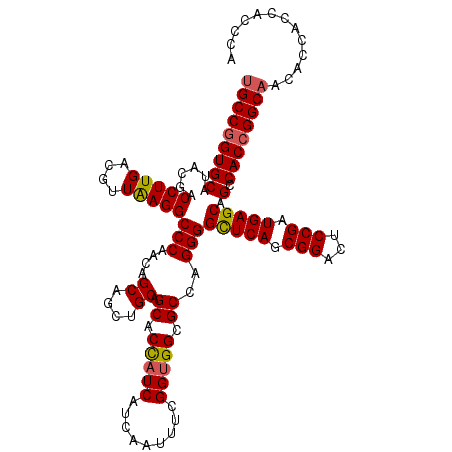

| Location | 22,058,540 – 22,058,660 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.90 |
| Mean single sequence MFE | -57.64 |
| Consensus MFE | -46.42 |
| Energy contribution | -47.10 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22058540 120 - 27905053 GGGCGCUGCGGUCGUGUGGGCGGUGGAGCAACUGCCACAGUAGGUGGUGGUGUUGCCGGUGGCUGCUCAUCGGAGUCCGCUGAGCCCCUGGCGCCACCGAAAUUGAUGAUGGUGCUGCAG .(((((((..((((......((((((((((.(..((((.....))))..)))))(((((.((..(((((.(((...))).)))))))))))).))))))....))))..))))))).... ( -55.50) >DroSec_CAF1 50616 120 - 1 GGGCGCUGCGGUCGAGUGGGCGGUGGAGCCACUGCCACAGUGGGUGGUGGUGUUGCCGGUGGCUGCUCAUCGGAGUCCGCUGAGCCCCUGGCGCCACCGAAAUUGAUGAUGGUGCUGCAG .(((((((..(((((.....((((((.(((((..((......))..)))))...(((((.((..(((((.(((...))).)))))))))))).))))))...)))))..))))))).... ( -59.70) >DroSim_CAF1 43300 120 - 1 GGGCGCUGCGGUCGUGUGGGCGGUGGAGCCACUGCCACAGUGGGGGGUGGUGUUGCCGGUGGCUGCUCAUCGGAGUCCGCUGAGCCCCUGGCGCCACCGAAAUUGAUGAUGGUGCUGCAG .(((((((..((((......((((((.((((((.((......)).))))))...(((((.((..(((((.(((...))).)))))))))))).))))))....))))..))))))).... ( -56.50) >DroEre_CAF1 45066 117 - 1 GGGCGCUGUGGUCGAGUGGGCGGUGGCGCGGUUG---CAGUGGGUGGUGGUGUGGCCGGUGGCUGCUCAUCGGAGUCCGCUGAACCCCUGGCGCCACCGAAAUUGAUGAUGGUGCUGCAG .((((((((.(((((.....(((((((((((((.---(((((((((((((.(..(((...)))..)))))))...)))))))))))....)))))))))...))))).)))))))).... ( -62.50) >DroYak_CAF1 52861 117 - 1 GGGCGCAGCGGUCGAGUGGGCGGUGGUGCAACUG---CAGUGGGUGGUGGUGUUGCCGGUGGCUGCUCAUCGGAGUCCGCUGAACCCCUGGCACCGCCGAAAUUGAUGAUAGUGCUGCAG ....(((((((((((.....(((((((((.....---(((.(((((((((.....(((((((....)))))))...)))))..))))))))))))))))...))))).....)))))).. ( -54.00) >consensus GGGCGCUGCGGUCGAGUGGGCGGUGGAGCAACUGCCACAGUGGGUGGUGGUGUUGCCGGUGGCUGCUCAUCGGAGUCCGCUGAGCCCCUGGCGCCACCGAAAUUGAUGAUGGUGCUGCAG .(((((((..(((((.....((((((.((........(((.(((((((((.....(((((((....)))))))...)))))..))))))))).))))))...)))))..))))))).... (-46.42 = -47.10 + 0.68)



| Location | 22,058,660 – 22,058,780 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -32.11 |
| Consensus MFE | -27.88 |
| Energy contribution | -28.48 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22058660 120 + 27905053 CAAGCUGCAGAGGGGCAUCACCAUCGUUGAGCACAUAGCUAUAACACCAACCAUGCCCCAAAAGUUCCAGAAUCGGCGCGCCAGGCUCUUCGACAUGUUCUUCCGCAGCUCCAAGAAACU ..((((((.(((((((((.......(((((((.....))).)))).......)))))))....(((..(((..(((....)).)..)))..))).......)).)))))).......... ( -35.34) >DroSec_CAF1 50736 120 + 1 CAAACUGCAGAGGGGCAUCACCAUUGUGGAGCACAUAGCUAUAACACCAACCAUGCCCCAAAAGUUCCAGAAUCGGCGCGCCAGGCUCUUCGACAUGUUCUUCCGCAGCUCCAAGAAACU ....((((.(((((((((......(((..(((.....)))...)))......)))))))....(((..(((..(((....)).)..)))..))).......)).))))............ ( -31.50) >DroSim_CAF1 43420 120 + 1 CAAACUGCAGAGGGGCAUCACCAUCGUGGAGCACAUAGCUAUAACACCAACCAUGCCCCAAAAGUUCCAGAAUCGGCGCGCCAGGCUCUUCGACAUGUUCUUCCGCAGCUCCAAGAAACU ....((((.(((((((((.......(((.(((.....)))....))).....)))))))....(((..(((..(((....)).)..)))..))).......)).))))............ ( -30.90) >DroEre_CAF1 45183 120 + 1 CAAGCUGCAGAGGGGCAUCACCAUUGUGGAGCACAUAGCUAUAACACCAACCACGCCCCAAAAGUUCCAGAAUCGGCGCGCCAGGCUCUUCGACAUGUUCUUCCGCAGCUCCAAGAAACU ..((((((.(((((((.((((....))))(((.....)))..............)))))....(((..(((..(((....)).)..)))..))).......)).)))))).......... ( -33.70) >DroYak_CAF1 52978 120 + 1 CAAACUGCAGCGGGGCAUCACCAUUGUGGAGCACAUUGCUAUAACACCAACCACGCCCCAAAAGUUCCAGAAUCGGCGUGCCAGGCUCUUCGACAUGUUCUUCCGCAGCUCCAAGAAACU ....((((...(((((.((((....))))(((.....)))..............)))))....(((..(((..(((....)).)..)))..)))..........))))............ ( -29.10) >consensus CAAACUGCAGAGGGGCAUCACCAUUGUGGAGCACAUAGCUAUAACACCAACCAUGCCCCAAAAGUUCCAGAAUCGGCGCGCCAGGCUCUUCGACAUGUUCUUCCGCAGCUCCAAGAAACU ....((((...(((((((.......(((.(((.....)))....))).....)))))))....(((..(((..(((....)).)..)))..)))..........))))............ (-27.88 = -28.48 + 0.60)

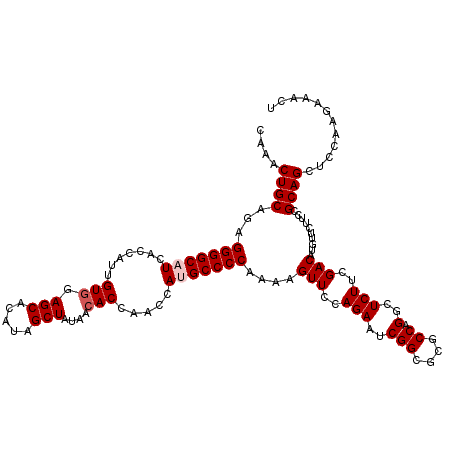
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:54 2006