
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,053,844 – 22,053,956 |
| Length | 112 |
| Max. P | 0.817922 |

| Location | 22,053,844 – 22,053,956 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.36 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -27.50 |
| Energy contribution | -26.45 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22053844 112 - 27905053 UUACUUUG-GGCCGAUCUAAAGAUCUCUCUGUGGAC----AUUAUCUAUGGCAUUUUAAAGUGUGAAAGCGAUCUUUUAGCGGCAUUUUAGGGGUGUGAAU---UUAACGCUCGUUUAAA ...((...-.((((....(((((((...(((((((.----....)))))))...........((....)))))))))...)))).....))((((((....---...))))))....... ( -28.30) >DroPse_CAF1 113089 119 - 1 UUGCUUUA-GGUUCAGUGAAAGAUCUCUCCGUAGAUACCUAUUAUCUACGGCACUUUAAAGUGUGAGAGCGAUCUUUUAGCGGCAUUUUAGGGGUGUGAAUUGUUUCACACUCGUUUCAA .((((...-.((....(((((((((.(((((((((((.....))))))))(((((....)))))..))).)))))))))))))))......(((((((((....)))))))))....... ( -40.60) >DroSec_CAF1 45963 112 - 1 UUACUUUG-GGCCGAUCUAGAGAUCUCUCUCUGGAC----AUUAUCUAUGGCAUUUUAAAGUGUGAAAGCGAUCUUUUAGCGGCAUUUUAGGGGUGUGAAU---UUAACGCUCGUUUUAA ...((...-.((((....(((((((.((.((((((.----....))))..(((((....))))))).)).)))))))...)))).....))((((((....---...))))))....... ( -27.90) >DroEre_CAF1 40372 112 - 1 UUGCUUUG-GGCCGAUCUAAAGAUCUCUCUGUGGAC----AUUAUCUAUGGCAUUUUAAAGUGCGAAAGCGAUCUUUUAGCGGCAUUUGAGGGGUGUGAAU---UUAACGCUCGUUUUAA ...(((..-.((((....(((((((...(((((((.----....)))))))...........((....)))))))))...))))....)))((((((....---...))))))....... ( -32.80) >DroYak_CAF1 48094 113 - 1 UUACUUUGGGGCCGAUCUAAAGAUCUCUCUGUGGAC----AUUAUCUAUGGCAUUUUAAAGUGUGAAAGCGAUCUUUUAGCGGCAUUUUAGGGGUGUGAAU---UUAACGCUCGUUUUAA ...((.....((((....(((((((...(((((((.----....)))))))...........((....)))))))))...)))).....))((((((....---...))))))....... ( -28.70) >DroPer_CAF1 132403 119 - 1 UUGCUUUA-GGUUCAGUGAAAGAUCUCUCCGUAGAUACCUAUUAUCUACGGCACUUUAAAGUGUGAGAGCGAUCUUUUAGCGGCAUUUUAGGGGUGUGAAUUGUUUCACACUCGUUUCAA .((((...-.((....(((((((((.(((((((((((.....))))))))(((((....)))))..))).)))))))))))))))......(((((((((....)))))))))....... ( -40.60) >consensus UUACUUUG_GGCCGAUCUAAAGAUCUCUCUGUGGAC____AUUAUCUAUGGCAUUUUAAAGUGUGAAAGCGAUCUUUUAGCGGCAUUUUAGGGGUGUGAAU___UUAACGCUCGUUUUAA ...((.....((((....(((((((...(((((((.........)))))))...........((....)))))))))...)))).....))((((((..........))))))....... (-27.50 = -26.45 + -1.05)
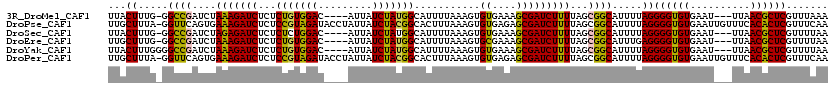
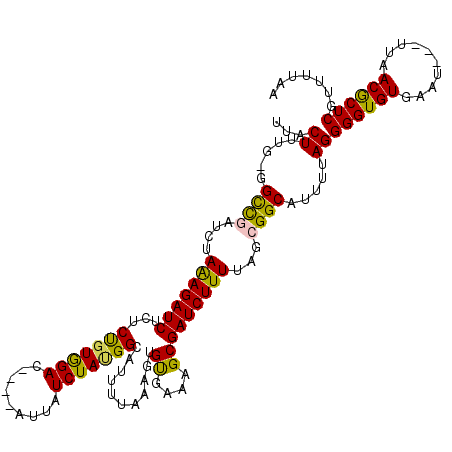

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:39 2006