
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,038,879 – 22,038,978 |
| Length | 99 |
| Max. P | 0.886501 |

| Location | 22,038,879 – 22,038,978 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -20.46 |
| Energy contribution | -21.68 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
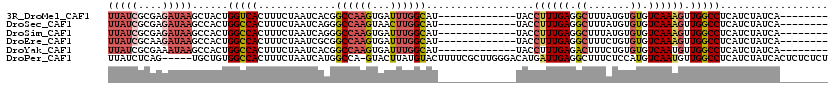
>3R_DroMel_CAF1 22038879 99 + 27905053 UUAUCGCGAGAUAAGCUACUGGUCACUUUCUAAUCACGGCCAAGUGAUUUGGCAU-------------UACCUUUGAGGCUUUAUGUGUGUCAAAGUUGGCCUCAUCUAUCA-------- (((((....)))))(((..(((((.............))))))))(((..(((..-------------...((((((.((.......)).))))))...)))..))).....-------- ( -25.92) >DroSec_CAF1 37089 99 + 1 UUAUCGCGAGAUAAGCCACUGGCCACUUUCUAAUCAGGGCCAAGUAACUUGGCAU-------------UACCUUUGAGGCUUUAUGUGUGUCAAAGUUGGCCUCAUCUAUCA-------- (((((....)))))......(((((.............((((((...))))))..-------------...((((((.((.......)).)))))).)))))..........-------- ( -30.10) >DroSim_CAF1 29512 99 + 1 UUAUCGCGAGAUAAGCCACUGGCCACUUUCUAAUCAGGGCCAAGUGAUUUGGCAU-------------UACCUUUGAGGCUUUAUGUGUGUCAAAGUUGGCCUCAUCUAUCA-------- (((((....)))))..((((((((.((........)))))).))))....(((..-------------...((((((.((.......)).))))))...)))..........-------- ( -30.40) >DroEre_CAF1 31040 99 + 1 UUAUCGCAAGAUAAGCCACUGGCCACUUUCUAAUCGCGGCCAAGUGAUUUGGCAU-------------UACCUUUGAGGCUUUCUGUGUGUCAAAGUUGGCCUCAUCUAUCA-------- (((((....)))))..((((((((.(.........).)))).))))....(((..-------------...((((((.((.......)).))))))...)))..........-------- ( -30.20) >DroYak_CAF1 38831 99 + 1 UUAUCGCGAAAUAAGCCACUGGCCACUUUCUAAUCACGGCCAAGUGAUUUGGCAU-------------UACCUUUGAGACUUUCUGUGUGUCAAUGUUGGCCUCAUCUAUCA-------- .....((.......))((((((((.............)))).))))....(((..-------------.((..((((.((.......)).)))).))..)))..........-------- ( -22.52) >DroPer_CAF1 122202 114 + 1 UUAUCUCAG-----UGCUGUGGCCACUUUCUAAUCAUGGCCA-GUACUUAUGUACUUUUCGCUUGGGACAUGAUUGAGGCUUUCUCCAUGUCAAUGUUGGCCUCAUCUAUCACUCUCUCU ...((((((-----.((..((((((...........))))))-((((....)))).....))))))))..(((((((((((.....(((....)))..)))))))...))))........ ( -31.70) >consensus UUAUCGCGAGAUAAGCCACUGGCCACUUUCUAAUCACGGCCAAGUGAUUUGGCAU_____________UACCUUUGAGGCUUUAUGUGUGUCAAAGUUGGCCUCAUCUAUCA________ (((((....)))))......(((((.............((((((...))))))..................((((((.((.......)).)))))).))))).................. (-20.46 = -21.68 + 1.22)
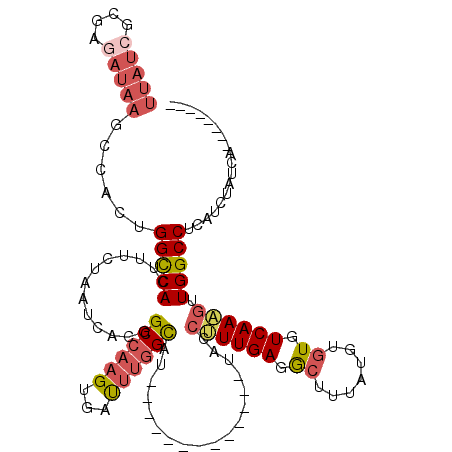
| Location | 22,038,879 – 22,038,978 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -26.33 |
| Consensus MFE | -17.23 |
| Energy contribution | -17.48 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
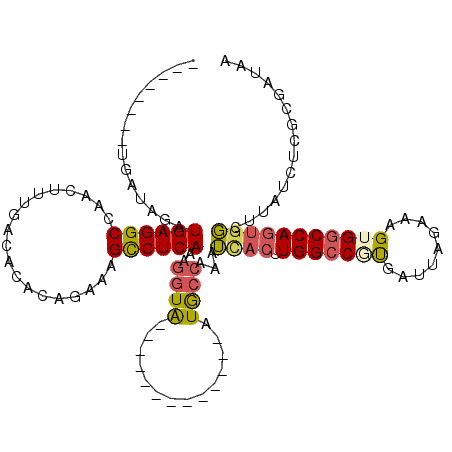
>3R_DroMel_CAF1 22038879 99 - 27905053 --------UGAUAGAUGAGGCCAACUUUGACACACAUAAAGCCUCAAAGGUA-------------AUGCCAAAUCACUUGGCCGUGAUUAGAAAGUGACCAGUAGCUUAUCUCGCGAUAA --------.......((((((......((.....))....))))))......-------------..(((((.....)))))(((((...((((((........)))).))))))).... ( -20.00) >DroSec_CAF1 37089 99 - 1 --------UGAUAGAUGAGGCCAACUUUGACACACAUAAAGCCUCAAAGGUA-------------AUGCCAAGUUACUUGGCCCUGAUUAGAAAGUGGCCAGUGGCUUAUCUCGCGAUAA --------.(..(((((((((...((((((..(.......)..))))))...-------------..)))..((((((.((((((........)).))))))))))))))))..)..... ( -28.20) >DroSim_CAF1 29512 99 - 1 --------UGAUAGAUGAGGCCAACUUUGACACACAUAAAGCCUCAAAGGUA-------------AUGCCAAAUCACUUGGCCCUGAUUAGAAAGUGGCCAGUGGCUUAUCUCGCGAUAA --------.......((((((......((.....))....)))))).(((((-------------(.((((......((((((((........)).))))))))))))))))........ ( -26.90) >DroEre_CAF1 31040 99 - 1 --------UGAUAGAUGAGGCCAACUUUGACACACAGAAAGCCUCAAAGGUA-------------AUGCCAAAUCACUUGGCCGCGAUUAGAAAGUGGCCAGUGGCUUAUCUUGCGAUAA --------.......((((((....((((.....))))..))))))((((((-------------(.((((......((((((((.........)))))))))))))))))))....... ( -32.40) >DroYak_CAF1 38831 99 - 1 --------UGAUAGAUGAGGCCAACAUUGACACACAGAAAGUCUCAAAGGUA-------------AUGCCAAAUCACUUGGCCGUGAUUAGAAAGUGGCCAGUGGCUUAUUUCGCGAUAA --------.....((..((((((...((((.((.......)).)))).((..-------------...)).......(((((((...........)))))))))))))...))....... ( -21.40) >DroPer_CAF1 122202 114 - 1 AGAGAGAGUGAUAGAUGAGGCCAACAUUGACAUGGAGAAAGCCUCAAUCAUGUCCCAAGCGAAAAGUACAUAAGUAC-UGGCCAUGAUUAGAAAGUGGCCACAGCA-----CUGAGAUAA .(.((..(((((...((((((...(((....)))......))))))))))).)).)..((....(((((....))))-)((((((.........))))))...)).-----......... ( -29.10) >consensus ________UGAUAGAUGAGGCCAACUUUGACACACAGAAAGCCUCAAAGGUA_____________AUGCCAAAUCACUUGGCCGUGAUUAGAAAGUGGCCAGUGGCUUAUCUCGCGAUAA ...............((((((...................))))))..((((..............))))...((((.(((((((.........)))))))))))............... (-17.23 = -17.48 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:30 2006