
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,037,630 – 22,037,799 |
| Length | 169 |
| Max. P | 0.999976 |

| Location | 22,037,630 – 22,037,720 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 90.68 |
| Mean single sequence MFE | -32.34 |
| Consensus MFE | -27.90 |
| Energy contribution | -27.42 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22037630 90 + 27905053 ACUUGAAGACACCAGCCACUCGAUUGCGGUGUUUU-----------GCGGGCAGGGGAUCCAUCGAGAUUGCAGCUCUCGGGAUCCACAUGCCAAACAUGU .....((((((((.((.........))))))))))-----------...((((..((((((..(((((.......)))))))))))...))))........ ( -32.70) >DroSec_CAF1 35879 90 + 1 ACUUGAAGCCAUCAGCCACUCCAUUGCGGUGUUUU-----------GUGGGCAGGGGAUCCAUCGAGAUUGCAGAUCUCGGGAUCCACAUGCCAAACAUGU .((((...((((..(((.(......).))).....-----------)))).))))((((((..(((((((...)))))))))))))(((((.....))))) ( -30.30) >DroSim_CAF1 28273 90 + 1 ACUUGAAGCCACCAGCCACUCGAUUGCGGUGUUUU-----------GUGGGCAGGGGAUCCAUCGAGAUUGCAGCUCUCGGGAUCCACAUGCCAAACAUGU .((((...((((.((.(((.((....))))).)).-----------)))).))))((((((..(((((.......)))))))))))(((((.....))))) ( -30.80) >DroEre_CAF1 29831 89 + 1 ACUUGAAGCCACCAGCCACCAGAUUGCGGUGUUUU------------GGGGCAGGGGGUCCAUCGAGAUUGCAGCUCUCGGGAUCCACAUGCCAAACAUGU .......(((.((((.((((.......))))..))------------)))))...((((((..(((((.......)))))))))))(((((.....))))) ( -31.60) >DroYak_CAF1 37566 101 + 1 ACUUGAAGCCACCAGCCACUCGAUUGUGGUGUUUUAGGGGGGGGGGGGGGGCAGGGGAUCCAUCGAGAUUGCAGCUCUCGGGAUCCACAUGCCAAACAUGU .((((((((.....(((((......))))))))))))).......(...((((..((((((..(((((.......)))))))))))...))))...).... ( -36.30) >consensus ACUUGAAGCCACCAGCCACUCGAUUGCGGUGUUUU___________GGGGGCAGGGGAUCCAUCGAGAUUGCAGCUCUCGGGAUCCACAUGCCAAACAUGU ....((((.((((.((.........))))))))))..............((((..((((((..(((((.......)))))))))))...))))........ (-27.90 = -27.42 + -0.48)



| Location | 22,037,630 – 22,037,720 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 90.68 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -28.18 |
| Energy contribution | -28.06 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22037630 90 - 27905053 ACAUGUUUGGCAUGUGGAUCCCGAGAGCUGCAAUCUCGAUGGAUCCCCUGCCCGC-----------AAAACACCGCAAUCGAGUGGCUGGUGUCUUCAAGU ...(((..((((.(.(((((((((((.......)))))..))))))).)))).))-----------)..(((((((.........)).)))))........ ( -30.30) >DroSec_CAF1 35879 90 - 1 ACAUGUUUGGCAUGUGGAUCCCGAGAUCUGCAAUCUCGAUGGAUCCCCUGCCCAC-----------AAAACACCGCAAUGGAGUGGCUGAUGGCUUCAAGU ...(((..((((.(.((((((((((((.....))))))..))))))).)))).))-----------)...((((((......)))).))............ ( -29.20) >DroSim_CAF1 28273 90 - 1 ACAUGUUUGGCAUGUGGAUCCCGAGAGCUGCAAUCUCGAUGGAUCCCCUGCCCAC-----------AAAACACCGCAAUCGAGUGGCUGGUGGCUUCAAGU ...(((..((((.(.(((((((((((.......)))))..))))))).)))).))-----------)...((((((.........)).))))......... ( -29.70) >DroEre_CAF1 29831 89 - 1 ACAUGUUUGGCAUGUGGAUCCCGAGAGCUGCAAUCUCGAUGGACCCCCUGCCCC------------AAAACACCGCAAUCUGGUGGCUGGUGGCUUCAAGU (((((.....)))))((.((((((((.......)))))..))).))...(((((------------(...(((((.....)))))..))).)))....... ( -29.20) >DroYak_CAF1 37566 101 - 1 ACAUGUUUGGCAUGUGGAUCCCGAGAGCUGCAAUCUCGAUGGAUCCCCUGCCCCCCCCCCCCCCCUAAAACACCACAAUCGAGUGGCUGGUGGCUUCAAGU .(((....((((.(.(((((((((((.......)))))..))))))).))))..................((((((......)))).)))))......... ( -29.50) >consensus ACAUGUUUGGCAUGUGGAUCCCGAGAGCUGCAAUCUCGAUGGAUCCCCUGCCCAC___________AAAACACCGCAAUCGAGUGGCUGGUGGCUUCAAGU .(((....((((.(.(((((((((((.......)))))..))))))).))))..................((((((......)))).)))))......... (-28.18 = -28.06 + -0.12)



| Location | 22,037,665 – 22,037,760 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 94.46 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -31.28 |
| Energy contribution | -31.12 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.94 |
| SVM decision value | 5.15 |
| SVM RNA-class probability | 0.999976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22037665 95 + 27905053 ------GCGGGCAGGGGAUCCAUCGAGAUUGCAGCUCUCGGGAUCCACAUGCCAAACAUGUUAUGCCUCGCAUAUAUAUUAAACGCUUAGUUAUAUGCUAC ------..(((((..((((((..(((((.......)))))))))))(((((.....)))))..))))).(((((((.(((((....))))))))))))... ( -32.80) >DroSec_CAF1 35914 95 + 1 ------GUGGGCAGGGGAUCCAUCGAGAUUGCAGAUCUCGGGAUCCACAUGCCAAACAUGUUAUGCCUCGCAUAUAUAUUAAACGCUUAGUUAUAUGCUAC ------..(((((..((((((..(((((((...)))))))))))))(((((.....)))))..))))).(((((((.(((((....))))))))))))... ( -34.50) >DroSim_CAF1 28308 95 + 1 ------GUGGGCAGGGGAUCCAUCGAGAUUGCAGCUCUCGGGAUCCACAUGCCAAACAUGUUAUGCCUCGCAUAUAUAUUAAACGCUUAGUUAUAUGCUAC ------..(((((..((((((..(((((.......)))))))))))(((((.....)))))..))))).(((((((.(((((....))))))))))))... ( -32.70) >DroEre_CAF1 29866 94 + 1 -------GGGGCAGGGGGUCCAUCGAGAUUGCAGCUCUCGGGAUCCACAUGCCAAACAUGUUAUACCUCGCAUAUAUAUUAAGCGCUUAGUUAUAUGCUAC -------..((((..((((((..(((((.......)))))))))))...))))................(((((((.(((((....))))))))))))... ( -31.20) >DroYak_CAF1 37606 101 + 1 GGGGGGGGGGGCAGGGGAUCCAUCGAGAUUGCAGCUCUCGGGAUCCACAUGCCAAACAUGUUAUACCUCGCAUAUAUAUUAAACGCUUAGUUAUAUGCUAC ((((.(...((((..((((((..(((((.......)))))))))))...))))...)........))))(((((((.(((((....))))))))))))... ( -35.00) >consensus ______GGGGGCAGGGGAUCCAUCGAGAUUGCAGCUCUCGGGAUCCACAUGCCAAACAUGUUAUGCCUCGCAUAUAUAUUAAACGCUUAGUUAUAUGCUAC .........((((..((((((..(((((.......)))))))))))...))))................(((((((.(((((....))))))))))))... (-31.28 = -31.12 + -0.16)



| Location | 22,037,665 – 22,037,760 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 94.46 |
| Mean single sequence MFE | -29.39 |
| Consensus MFE | -29.08 |
| Energy contribution | -29.04 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.14 |
| SVM RNA-class probability | 0.999813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22037665 95 - 27905053 GUAGCAUAUAACUAAGCGUUUAAUAUAUAUGCGAGGCAUAACAUGUUUGGCAUGUGGAUCCCGAGAGCUGCAAUCUCGAUGGAUCCCCUGCCCGC------ ...(((((((...............)))))))(.((((..(((((.....)))))(((((((((((.......)))))..))))))..)))))..------ ( -31.06) >DroSec_CAF1 35914 95 - 1 GUAGCAUAUAACUAAGCGUUUAAUAUAUAUGCGAGGCAUAACAUGUUUGGCAUGUGGAUCCCGAGAUCUGCAAUCUCGAUGGAUCCCCUGCCCAC------ ...(((((((...............)))))))(.((((..(((((.....)))))((((((((((((.....))))))..))))))..)))))..------ ( -31.56) >DroSim_CAF1 28308 95 - 1 GUAGCAUAUAACUAAGCGUUUAAUAUAUAUGCGAGGCAUAACAUGUUUGGCAUGUGGAUCCCGAGAGCUGCAAUCUCGAUGGAUCCCCUGCCCAC------ ...(((((((...............)))))))(.((((..(((((.....)))))(((((((((((.......)))))..))))))..)))))..------ ( -31.06) >DroEre_CAF1 29866 94 - 1 GUAGCAUAUAACUAAGCGCUUAAUAUAUAUGCGAGGUAUAACAUGUUUGGCAUGUGGAUCCCGAGAGCUGCAAUCUCGAUGGACCCCCUGCCCC------- ...(((((((..(((....)))...)))))))..((((..(((((.....)))))((.((((((((.......)))))..))).))..))))..------- ( -24.70) >DroYak_CAF1 37606 101 - 1 GUAGCAUAUAACUAAGCGUUUAAUAUAUAUGCGAGGUAUAACAUGUUUGGCAUGUGGAUCCCGAGAGCUGCAAUCUCGAUGGAUCCCCUGCCCCCCCCCCC ...(((((((...............)))))))..((((..(((((.....)))))(((((((((((.......)))))..))))))..))))......... ( -28.56) >consensus GUAGCAUAUAACUAAGCGUUUAAUAUAUAUGCGAGGCAUAACAUGUUUGGCAUGUGGAUCCCGAGAGCUGCAAUCUCGAUGGAUCCCCUGCCCAC______ ...(((((((...............)))))))(.((((..(((((.....)))))(((((((((((.......)))))..))))))..)))))........ (-29.08 = -29.04 + -0.04)

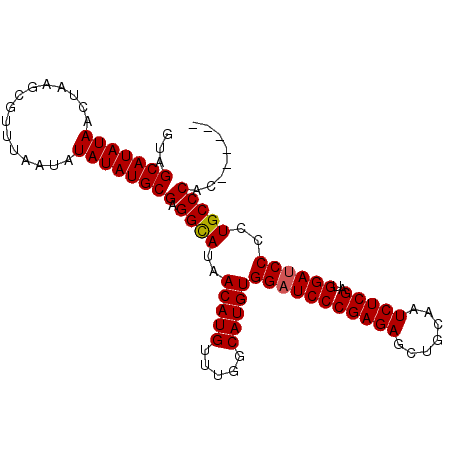

| Location | 22,037,696 – 22,037,799 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -10.27 |
| Energy contribution | -12.17 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22037696 103 + 27905053 UCGGGAUCCACAUGCC----------------AAACAUGUUAUGCCUCGCAUAUAUAUUAAACGCUUAGUUAUAUGCUACUUAACACUUCCCUCCAUUUAUUUUUGCUCGAGAGCGAAA ..((((...(((((..----------------...)))))........(((((((.(((((....))))))))))))...........)))).........(((((((....))))))) ( -22.40) >DroPse_CAF1 101476 112 + 1 UCA-UAUCCGCAUGCUCCCUCUACGAGUAUUCAUAUACAU-AUGUA-UGUACAUAUAUUAAACGCUUAGUUAUAUGCUACUUAACACUUCUCUUCA----UUUUCGCUGGAGAGCGAAA ...-........(((((..(((((((((((....))))..-.(((.-.(((((((((((((....)))).)))))).)))...)))..........----...))).)))))))))... ( -19.30) >DroSec_CAF1 35945 103 + 1 UCGGGAUCCACAUGCC----------------AAACAUGUUAUGCCUCGCAUAUAUAUUAAACGCUUAGUUAUAUGCUACUUAACACUUCCCUCCAUUUAUUUUUGCUCGAGAGCGAAA ..((((...(((((..----------------...)))))........(((((((.(((((....))))))))))))...........)))).........(((((((....))))))) ( -22.40) >DroSim_CAF1 28339 103 + 1 UCGGGAUCCACAUGCC----------------AAACAUGUUAUGCCUCGCAUAUAUAUUAAACGCUUAGUUAUAUGCUACUUAACACUUCCCUCCAUUUAUUUUUUCUCGAGAGCGAAA ..((((...(((((..----------------...)))))........(((((((.(((((....))))))))))))...........))))...........((((.(....).)))) ( -19.60) >DroEre_CAF1 29896 103 + 1 UCGGGAUCCACAUGCC----------------AAACAUGUUAUACCUCGCAUAUAUAUUAAGCGCUUAGUUAUAUGCUACUUAACACUUCCCUCCAUUUAUUCUUGCUCUGGAGCGAAA ..((((...(((((..----------------...)))))........(((((((.(((((....))))))))))))...........))))...........(((((....))))).. ( -22.00) >DroPer_CAF1 120814 107 + 1 UCA-UAUCCGCAUGCUCUCUCUACGAGUAUUCAUAUAUGU-AUGUA-U----A-AUAUUAAACGCUUAGUUAUAUGCUACUUAACACUUCUCUUCA----UUUUCGCUGGAGAGCGAAA ...-........(((((((.(..(((((((....))))((-(.(((-(----(-..(((((....)))))..))))))))................----...)))..))))))))... ( -18.90) >consensus UCGGGAUCCACAUGCC________________AAACAUGUUAUGCCUCGCAUAUAUAUUAAACGCUUAGUUAUAUGCUACUUAACACUUCCCUCCAUUUAUUUUUGCUCGAGAGCGAAA ..((((...(((((.....................)))))........(((((((.(((((....))))))))))))...........)))).........(((((((....))))))) (-10.27 = -12.17 + 1.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:28 2006