
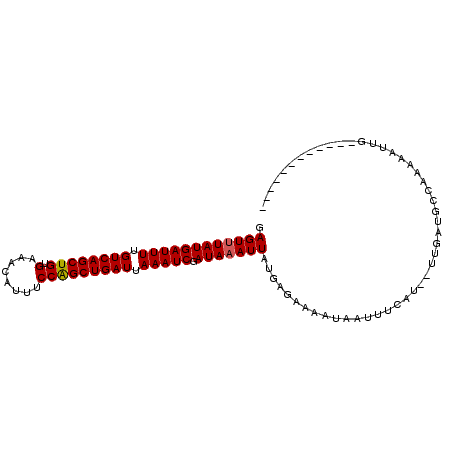
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,822,413 – 2,822,506 |
| Length | 93 |
| Max. P | 0.959814 |

| Location | 2,822,413 – 2,822,506 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.13 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.58 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
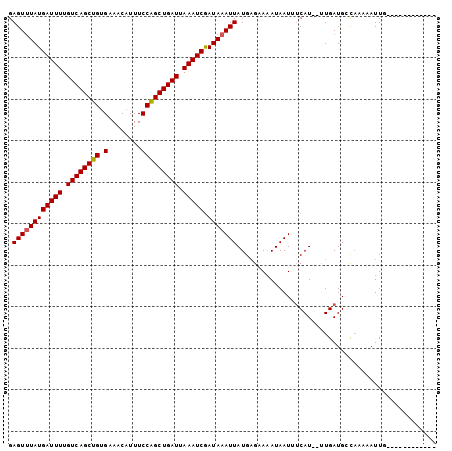
>3R_DroMel_CAF1 2822413 93 + 27905053 GAGUUUAUGAUUUUGUCAGCUGUGUAACAUUUCCAGCUGAUAAAAUCAAUAAAUUAUGAGAAAAUAAUUUCAU--UUGAUGCCAUCUAUUUGAUU-------- .(((((((((((((((((((((.(........))))))))))))))).)))))))((((((......))))))--....................-------- ( -25.50) >DroVir_CAF1 156248 96 + 1 GAGUUUAUGAUUUUGUCAGCUGUGAAACAUUUCCAGCUGAUUAAAUCGAUAAAUUAUGAGAAUAUAAUAACAU--CUGAUGACAAAAAUUG----GUAAA-AU .((((((((((((.((((((((.(((....))))))))))).))))).)))))))..................--(..((.......))..----)....-.. ( -22.60) >DroGri_CAF1 154151 96 + 1 GAGUUUAUGAUUUUGUCAGCUGUGAAACAUUUCCAGCUGAUUAAAUCGAUAAAUUAUGUGCAAAUAAUGUCAU--UUGAUGAUUAAAAUUG----GUCGA-AU .((((((((((((.((((((((.(((....))))))))))).))))).)))))))................((--(((((.(.......).----)))))-)) ( -25.70) >DroSim_CAF1 102216 93 + 1 GAGUUUAUGAUUUUGUCAGCUGCGUAACAUUUCCAGCUGAUUAAAUCGAUAAAUUAUGAGAAAAUAAUUUCAU--UUGAUGCCAACUAUUUGAUU-------- .((((((((((((.((((((((.(........))))))))).))))).)))))))((((((......))))))--....................-------- ( -21.90) >DroYak_CAF1 100944 93 + 1 GAGUAUAUGAUUUUGUCAGCUGUGUAACAUUUCCAGCUGAUUAAAUCGAUAAAUUAUGAGAAAAUAAUUUCAU--UUGAUGCCAACAAUUUGAUU-------- (.((((.((((((.((((((((.(........))))))))).)))))).......((((((......))))))--...)))))............-------- ( -20.60) >DroMoj_CAF1 162605 99 + 1 GAGUUUAUGAUUUUGUCAGCUGUGAAACAUUUCCGGCUGAUUAAAUCGAUAAAUUAUGAGCAAAUAAUAUCAUUGCUGAUGGCAAAAAUUG----UUAAAAAC .((((((((((((.((((((((.(((....))))))))))).))))).))))))).(.(((((.........))))).)(((((.....))----)))..... ( -25.70) >consensus GAGUUUAUGAUUUUGUCAGCUGUGAAACAUUUCCAGCUGAUUAAAUCGAUAAAUUAUGAGAAAAUAAUUUCAU__UUGAUGCCAAAAAUUG____________ .((((((((((((.((((((((.(........))))))))).))))).)))))))................................................ (-19.56 = -19.58 + 0.03)

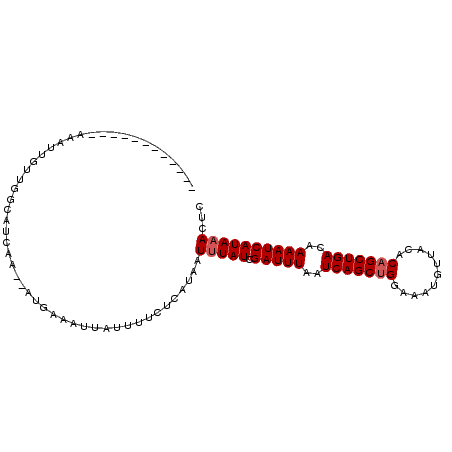
| Location | 2,822,413 – 2,822,506 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.13 |
| Mean single sequence MFE | -17.72 |
| Consensus MFE | -14.18 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
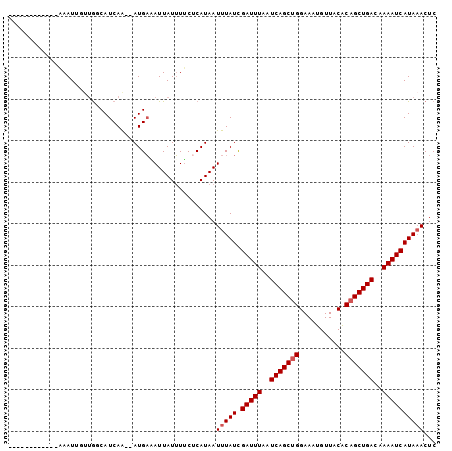
>3R_DroMel_CAF1 2822413 93 - 27905053 --------AAUCAAAUAGAUGGCAUCAA--AUGAAAUUAUUUUCUCAUAAUUUAUUGAUUUUAUCAGCUGGAAAUGUUACACAGCUGACAAAAUCAUAAACUC --------....................--...(((((((......)))))))..(((((((.(((((((...........))))))).)))))))....... ( -18.40) >DroVir_CAF1 156248 96 - 1 AU-UUUAC----CAAUUUUUGUCAUCAG--AUGUUAUUAUAUUCUCAUAAUUUAUCGAUUUAAUCAGCUGGAAAUGUUUCACAGCUGACAAAAUCAUAAACUC ..-.....----...............(--(((..(((((......))))).))))(((((..((((((((((....))).)))))))..)))))........ ( -19.50) >DroGri_CAF1 154151 96 - 1 AU-UCGAC----CAAUUUUAAUCAUCAA--AUGACAUUAUUUGCACAUAAUUUAUCGAUUUAAUCAGCUGGAAAUGUUUCACAGCUGACAAAAUCAUAAACUC ..-.....----.............(((--(((....)))))).......(((((.(((((..((((((((((....))).)))))))..))))))))))... ( -18.50) >DroSim_CAF1 102216 93 - 1 --------AAUCAAAUAGUUGGCAUCAA--AUGAAAUUAUUUUCUCAUAAUUUAUCGAUUUAAUCAGCUGGAAAUGUUACGCAGCUGACAAAAUCAUAAACUC --------....((((((((..(((...--))).))))))))........(((((.(((((..((((((((........).)))))))..))))))))))... ( -17.60) >DroYak_CAF1 100944 93 - 1 --------AAUCAAAUUGUUGGCAUCAA--AUGAAAUUAUUUUCUCAUAAUUUAUCGAUUUAAUCAGCUGGAAAUGUUACACAGCUGACAAAAUCAUAUACUC --------....(((((((.((....((--(((....))))).)).)))))))...(((((..(((((((...........)))))))..)))))........ ( -16.50) >DroMoj_CAF1 162605 99 - 1 GUUUUUAA----CAAUUUUUGCCAUCAGCAAUGAUAUUAUUUGCUCAUAAUUUAUCGAUUUAAUCAGCCGGAAAUGUUUCACAGCUGACAAAAUCAUAAACUC ........----...........((((....))))...............(((((.(((((..(((((.((((....))).).)))))..))))))))))... ( -15.80) >consensus ____________AAAUUGUUGGCAUCAA__AUGAAAUUAUUUUCUCAUAAUUUAUCGAUUUAAUCAGCUGGAAAUGUUACACAGCUGACAAAAUCAUAAACUC ..................................................(((((.(((((..(((((((...........)))))))..))))))))))... (-14.18 = -14.52 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:58 2006