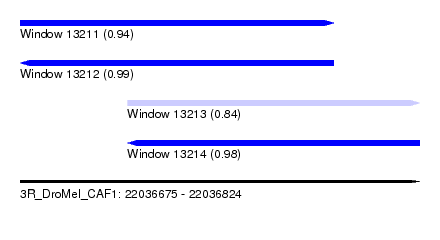
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,036,675 – 22,036,824 |
| Length | 149 |
| Max. P | 0.990053 |

| Location | 22,036,675 – 22,036,792 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 94.79 |
| Mean single sequence MFE | -38.70 |
| Consensus MFE | -34.98 |
| Energy contribution | -36.14 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22036675 117 + 27905053 UCGGCUCAAAUUGCAACUUUUGAUUAGCACUCUGAUGUAAUAAUCAACUUUGGGCCACAAAUCCUGCCGUGGCAUUUGCUGGGCAGGAAAAGCGGUGCGGCUGGCAUUUUGUGGGGC ..((((((((.........(((((((..((......))..))))))).))))))))((((((((((((.((((....)))))))))))......((((.....)))))))))..... ( -37.40) >DroSec_CAF1 34912 117 + 1 UCGGCUCAAAUUGCAACUUUUGAUUAGCACUCUGAUGUAAUAAUCAACUUUGGGCCACAAAUCCUGCCGUGGCAUUUGCUGGGCAGGAAAAGCGGUGCGGCUGGCAUUGUGUGGGGC ..((((((((.........(((((((..((......))..))))))).)))))))).....(((((((.((((....)))))))))))...(((((((.....)))))))....... ( -40.00) >DroSim_CAF1 27203 117 + 1 UCGGCUCAAAUUGCAACUUUUGAUUAGCACUCUGAUGUAAUAAUCAACUUUGGGCCACAAAUCCUGCCGUGGCAUUUGCUGGGCAGGAAAAGCGGUGCGGCUGGCAUUGAGUGGGGC ((.((((((..(((.....(((((((..((......))..))))))).....((((.....(((((((.((((....)))))))))))...((...)))))).))))))))).)).. ( -41.10) >DroEre_CAF1 28931 117 + 1 UCGGCCCAAAUUGCAACUUUUGAUUAGCACUCUGAUGUAAUAAUCAACUUUGGGCCACAAAUCCUGCCAUGGCAUUUGCCGGGCAGGAAAAGCGUUGCUGUUGGCAUUGUGAGGGGC ...((((...(..(((...(((((((..((......))..)))))))......((((....(((((((.((((....)))))))))))..(((...)))..)))).)))..).)))) ( -43.80) >DroYak_CAF1 36606 117 + 1 UCGGCACAAAUUGCAACUUUUGAUUAGCACUCUGAUGUAAUAAUCAACUUUGGGCCACAAAUCCUGCCGUCGAAUUUGCUGGGCAGGAAAAGCGGUACGGCUGGCAUUGUGAGUGGU (((.(((((....(((...(((((((..((......))..)))))))..))).(((.....(((((((.............)))))))..(((......)))))).)))))..))). ( -31.22) >consensus UCGGCUCAAAUUGCAACUUUUGAUUAGCACUCUGAUGUAAUAAUCAACUUUGGGCCACAAAUCCUGCCGUGGCAUUUGCUGGGCAGGAAAAGCGGUGCGGCUGGCAUUGUGUGGGGC ..((((((((.........(((((((..((......))..))))))).)))))))).....(((((((.((((....)))))))))))...(((((((.....)))))))....... (-34.98 = -36.14 + 1.16)


| Location | 22,036,675 – 22,036,792 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 94.79 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -29.88 |
| Energy contribution | -29.96 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.990053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22036675 117 - 27905053 GCCCCACAAAAUGCCAGCCGCACCGCUUUUCCUGCCCAGCAAAUGCCACGGCAGGAUUUGUGGCCCAAAGUUGAUUAUUACAUCAGAGUGCUAAUCAAAAGUUGCAAUUUGAGCCGA ......((((.(((.(((.(..((((...(((((((..((....))...)))))))...))))..)....(((((((.(((......))).)))))))..)))))).))))...... ( -34.70) >DroSec_CAF1 34912 117 - 1 GCCCCACACAAUGCCAGCCGCACCGCUUUUCCUGCCCAGCAAAUGCCACGGCAGGAUUUGUGGCCCAAAGUUGAUUAUUACAUCAGAGUGCUAAUCAAAAGUUGCAAUUUGAGCCGA ............((((((((((.......(((((((..((....))...)))))))..)))))).(((..(((((((.(((......))).)))))))...))).....)).))... ( -32.20) >DroSim_CAF1 27203 117 - 1 GCCCCACUCAAUGCCAGCCGCACCGCUUUUCCUGCCCAGCAAAUGCCACGGCAGGAUUUGUGGCCCAAAGUUGAUUAUUACAUCAGAGUGCUAAUCAAAAGUUGCAAUUUGAGCCGA ......((((((((.(((.(..((((...(((((((..((....))...)))))))...))))..)....(((((((.(((......))).)))))))..))))))..))))).... ( -35.50) >DroEre_CAF1 28931 117 - 1 GCCCCUCACAAUGCCAACAGCAACGCUUUUCCUGCCCGGCAAAUGCCAUGGCAGGAUUUGUGGCCCAAAGUUGAUUAUUACAUCAGAGUGCUAAUCAAAAGUUGCAAUUUGGGCCGA ...........(((.....)))..((...(((((((.(((....)))..)))))))...))((((((((.(((((((.(((......))).))))))).........)))))))).. ( -41.00) >DroYak_CAF1 36606 117 - 1 ACCACUCACAAUGCCAGCCGUACCGCUUUUCCUGCCCAGCAAAUUCGACGGCAGGAUUUGUGGCCCAAAGUUGAUUAUUACAUCAGAGUGCUAAUCAAAAGUUGCAAUUUGUGCCGA ......((((((((.(((.(..((((...(((((((...(......)..)))))))...))))..)....(((((((.(((......))).)))))))..))))))..))))).... ( -33.50) >consensus GCCCCACACAAUGCCAGCCGCACCGCUUUUCCUGCCCAGCAAAUGCCACGGCAGGAUUUGUGGCCCAAAGUUGAUUAUUACAUCAGAGUGCUAAUCAAAAGUUGCAAUUUGAGCCGA ............(((((..(((((((...(((((((..((....))...)))))))...)))).......(((((((.(((......))).)))))))....)))...))).))... (-29.88 = -29.96 + 0.08)

| Location | 22,036,715 – 22,036,824 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -26.72 |
| Energy contribution | -27.32 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.836791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22036715 109 + 27905053 UAAUCAACUUUGGGCCACAAAUCCUGCCGUGGCAUUUGCUGGGCAGGAAAAGCGGUGCGGCUGGCAUUUUGUGGGGC-GUUCAUAAAUACAUAUUAUUGACUGAAUGAAG ..............((((((((((((((.((((....)))))))))))......((((.....)))))))))))..(-(((((..((((.....))))...))))))... ( -33.30) >DroSec_CAF1 34952 109 + 1 UAAUCAACUUUGGGCCACAAAUCCUGCCGUGGCAUUUGCUGGGCAGGAAAAGCGGUGCGGCUGGCAUUGUGUGGGGC-AUUCAUAAAUACAUAUUAUUGACUGAAUAAAG .......(((((..((((...(((((((.((((....)))))))))))...(((((((.....)))))))))))...-.((((..((((.....))))...))))))))) ( -33.40) >DroSim_CAF1 27243 109 + 1 UAAUCAACUUUGGGCCACAAAUCCUGCCGUGGCAUUUGCUGGGCAGGAAAAGCGGUGCGGCUGGCAUUGAGUGGGGC-AUUCAUAAAUACAUAUUAUUGACUGAAUGAAG ..............((((...(((((((.((((....)))))))))))....((((((.....)))))).))))..(-(((((..((((.....))))...))))))... ( -34.60) >DroEre_CAF1 28971 110 + 1 UAAUCAACUUUGGGCCACAAAUCCUGCCAUGGCAUUUGCCGGGCAGGAAAAGCGUUGCUGUUGGCAUUGUGAGGGGCAAUUCAUAAAUACAUAUUAUUGAGUGAAUGAAG .......((((..(((.....(((((((.((((....)))))))))))...(((.(((.....))).)))....))).((((((.((((.....))))..)))))))))) ( -34.10) >DroYak_CAF1 36646 109 + 1 UAAUCAACUUUGGGCCACAAAUCCUGCCGUCGAAUUUGCUGGGCAGGAAAAGCGGUACGGCUGGCAUUGUGAGUGGU-AUUCAUAAAUACAUAUUAUUCACUGAAUGAAG ...(((.......(((.....(((((((.............)))))))..(((......))))))...(((((((((-((..........)))))))))))....))).. ( -29.62) >consensus UAAUCAACUUUGGGCCACAAAUCCUGCCGUGGCAUUUGCUGGGCAGGAAAAGCGGUGCGGCUGGCAUUGUGUGGGGC_AUUCAUAAAUACAUAUUAUUGACUGAAUGAAG .......((((..(((.....(((((((.((((....)))))))))))...(((((((.....)))))))....))).(((((..((((.....))))...))))))))) (-26.72 = -27.32 + 0.60)

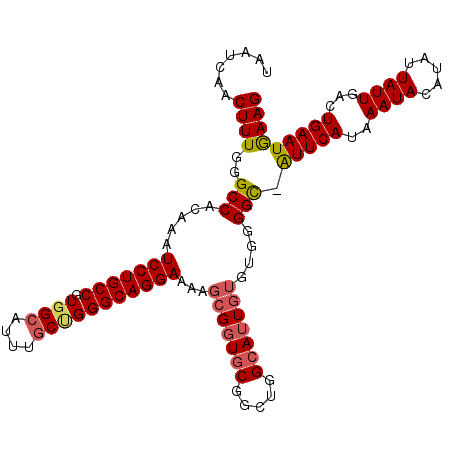
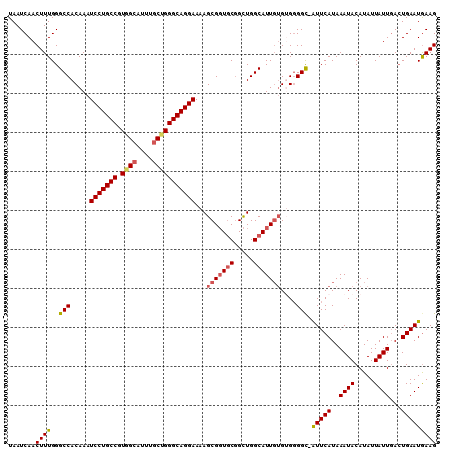
| Location | 22,036,715 – 22,036,824 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 93.24 |
| Mean single sequence MFE | -27.82 |
| Consensus MFE | -22.98 |
| Energy contribution | -23.62 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.976892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22036715 109 - 27905053 CUUCAUUCAGUCAAUAAUAUGUAUUUAUGAAC-GCCCCACAAAAUGCCAGCCGCACCGCUUUUCCUGCCCAGCAAAUGCCACGGCAGGAUUUGUGGCCCAAAGUUGAUUA ........(((((((.....((((((.((...-....))..))))))..((((((.......(((((((..((....))...)))))))..)))))).....))))))). ( -28.50) >DroSec_CAF1 34952 109 - 1 CUUUAUUCAGUCAAUAAUAUGUAUUUAUGAAU-GCCCCACACAAUGCCAGCCGCACCGCUUUUCCUGCCCAGCAAAUGCCACGGCAGGAUUUGUGGCCCAAAGUUGAUUA ........(((((((.....(((((....)))-))..............((((((.......(((((((..((....))...)))))))..)))))).....))))))). ( -28.40) >DroSim_CAF1 27243 109 - 1 CUUCAUUCAGUCAAUAAUAUGUAUUUAUGAAU-GCCCCACUCAAUGCCAGCCGCACCGCUUUUCCUGCCCAGCAAAUGCCACGGCAGGAUUUGUGGCCCAAAGUUGAUUA ...((((((...((((.....))))..)))))-)......(((((....((((((.......(((((((..((....))...)))))))..)))))).....)))))... ( -30.00) >DroEre_CAF1 28971 110 - 1 CUUCAUUCACUCAAUAAUAUGUAUUUAUGAAUUGCCCCUCACAAUGCCAACAGCAACGCUUUUCCUGCCCGGCAAAUGCCAUGGCAGGAUUUGUGGCCCAAAGUUGAUUA ..........(((((..((((....))))....(((........(((.....)))..((...(((((((.(((....)))..)))))))...))))).....)))))... ( -26.30) >DroYak_CAF1 36646 109 - 1 CUUCAUUCAGUGAAUAAUAUGUAUUUAUGAAU-ACCACUCACAAUGCCAGCCGUACCGCUUUUCCUGCCCAGCAAAUUCGACGGCAGGAUUUGUGGCCCAAAGUUGAUUA ....(((((.((((((.....)))))))))))-..............((((.(..((((...(((((((...(......)..)))))))...))))..)...)))).... ( -25.90) >consensus CUUCAUUCAGUCAAUAAUAUGUAUUUAUGAAU_GCCCCACACAAUGCCAGCCGCACCGCUUUUCCUGCCCAGCAAAUGCCACGGCAGGAUUUGUGGCCCAAAGUUGAUUA ........(((((((.............................(((.....)))((((...(((((((..((....))...)))))))...))))......))))))). (-22.98 = -23.62 + 0.64)

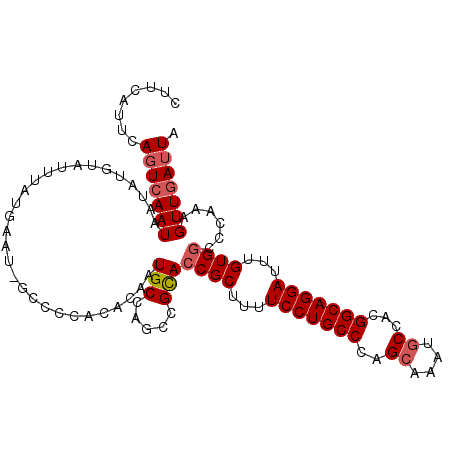
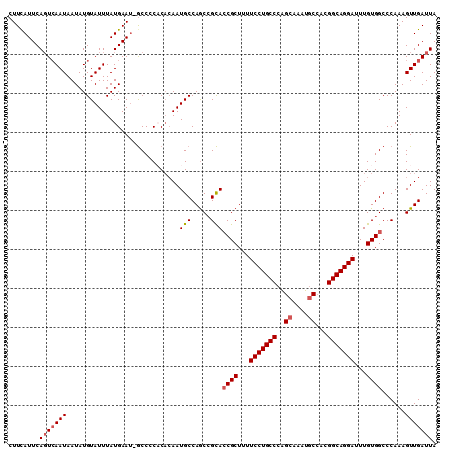
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:24 2006