| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,035,901 – 22,036,025 |
| Length | 124 |
| Max. P | 0.997497 |

| Location | 22,035,901 – 22,035,998 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 96.29 |
| Mean single sequence MFE | -21.48 |
| Consensus MFE | -19.10 |
| Energy contribution | -21.00 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22035901 97 + 27905053 UCAUUGAGAAGGAAAGCGAGGGGAUCCUGCCAGAAGGGUAAGCAAUUGCCAACAAUUCCCAAUAGGAAACGAAAAUGGUAGGAAUAGAAUAACGUAA ........................((((((((....(((((....))))).....((((.....)))).......)))))))).............. ( -22.30) >DroSec_CAF1 34137 97 + 1 UCAUUGAGAAGGAGAGCGAGGGGAUCCUGCCAGAAGGGUAAGCAAUUGCCAACAAUUCCCAAUAGGAAACGAAAAUGGUAGGAAUAGAAUAACGUAA ........................((((((((....(((((....))))).....((((.....)))).......)))))))).............. ( -22.30) >DroSim_CAF1 26429 97 + 1 UCAUUGAGAAGGAGAGCGAGGGGAUCCUGCCAGAAGGGUAAGCAAUUGCCAACAAUUCCCAAUAGGAAACGAAAAUGGUAGGAAUAGAAUAACGUAA ........................((((((((....(((((....))))).....((((.....)))).......)))))))).............. ( -22.30) >DroEre_CAF1 28206 95 + 1 UCAUUGAGGAGGAAAACGAGGGGAUCCUGC--GAAGGGUAAGCAAUUGCCAACAAUUCCCAAUAGGAAACGAAAAUGGUAGGAAUAAAAUAACGUAA ((((((.((((.....(((((....))).)--)...(((((....))))).....)))))))).(....)))......................... ( -18.20) >DroYak_CAF1 35836 97 + 1 UCAUUGAGAAGGAAAACGAGGGGAUCCUGCCAGAAGGGUAAGCAAUUGCCAACAAUUCCCAAUAGGAAACGAAAAUGGUAGGAAUAGGAUAACGCAA ........................((((((((....(((((....))))).....((((.....)))).......)))))))).............. ( -22.30) >consensus UCAUUGAGAAGGAAAGCGAGGGGAUCCUGCCAGAAGGGUAAGCAAUUGCCAACAAUUCCCAAUAGGAAACGAAAAUGGUAGGAAUAGAAUAACGUAA ........................((((((((....(((((....))))).....((((.....)))).......)))))))).............. (-19.10 = -21.00 + 1.90)


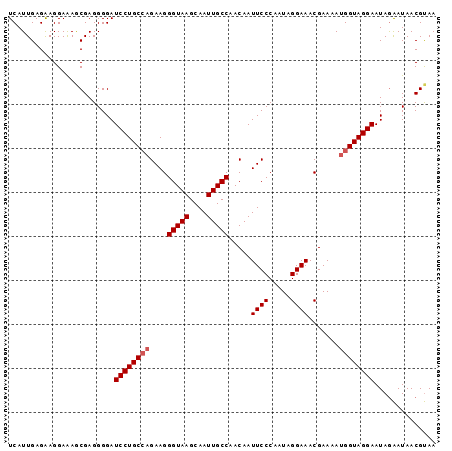
| Location | 22,035,922 – 22,036,025 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 97.09 |
| Mean single sequence MFE | -27.36 |
| Consensus MFE | -23.20 |
| Energy contribution | -25.30 |
| Covariance contribution | 2.10 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22035922 103 + 27905053 GGAUCCUGCCAGAAGGGUAAGCAAUUGCCAACAAUUCCCAAUAGGAAACGAAAAUGGUAGGAAUAGAAUAACGUAAAAGGUCAAUAGGCCAAUGGAUACUCUA (((((((((((....(((((....))))).....((((.....)))).......))))))))................((((....)))).........))). ( -27.70) >DroSec_CAF1 34158 103 + 1 GGAUCCUGCCAGAAGGGUAAGCAAUUGCCAACAAUUCCCAAUAGGAAACGAAAAUGGUAGGAAUAGAAUAACGUAAAAGGUCAAUAGGCCAAUGGAUACUCUA (((((((((((....(((((....))))).....((((.....)))).......))))))))................((((....)))).........))). ( -27.70) >DroSim_CAF1 26450 103 + 1 GGAUCCUGCCAGAAGGGUAAGCAAUUGCCAACAAUUCCCAAUAGGAAACGAAAAUGGUAGGAAUAGAAUAACGUAAAAGGUCAAUAGGCCAAUGGAUACUCUA (((((((((((....(((((....))))).....((((.....)))).......))))))))................((((....)))).........))). ( -27.70) >DroEre_CAF1 28227 101 + 1 GGAUCCUGC--GAAGGGUAAGCAAUUGCCAACAAUUCCCAAUAGGAAACGAAAAUGGUAGGAAUAAAAUAACGUAAAAGGGCAAUAGGCCAAUGGAUACCCUA (((((((((--(...(((((....)))))....(((((((...(....).....))...))))).......))))....(((.....)))...)))).))... ( -24.00) >DroYak_CAF1 35857 103 + 1 GGAUCCUGCCAGAAGGGUAAGCAAUUGCCAACAAUUCCCAAUAGGAAACGAAAAUGGUAGGAAUAGGAUAACGCAAAAGGUCAAUAGGCCAAUGGAUACCCUA ...((((((((....(((((....))))).....((((.....)))).......)))))))).((((.((.(.((...((((....))))..))).)).)))) ( -29.70) >consensus GGAUCCUGCCAGAAGGGUAAGCAAUUGCCAACAAUUCCCAAUAGGAAACGAAAAUGGUAGGAAUAGAAUAACGUAAAAGGUCAAUAGGCCAAUGGAUACUCUA ...((((((((....(((((....))))).....((((.....)))).......))))))))................((((....))))............. (-23.20 = -25.30 + 2.10)


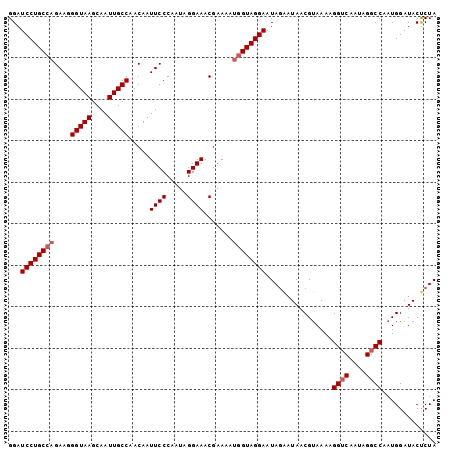
| Location | 22,035,922 – 22,036,025 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 97.09 |
| Mean single sequence MFE | -21.44 |
| Consensus MFE | -19.64 |
| Energy contribution | -19.48 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22035922 103 - 27905053 UAGAGUAUCCAUUGGCCUAUUGACCUUUUACGUUAUUCUAUUCCUACCAUUUUCGUUUCCUAUUGGGAAUUGUUGGCAAUUGCUUACCCUUCUGGCAGGAUCC (((((((......((.(....).))........))))))).((((.(((.....((((((....)))))).((.(((....))).)).....))).))))... ( -21.04) >DroSec_CAF1 34158 103 - 1 UAGAGUAUCCAUUGGCCUAUUGACCUUUUACGUUAUUCUAUUCCUACCAUUUUCGUUUCCUAUUGGGAAUUGUUGGCAAUUGCUUACCCUUCUGGCAGGAUCC (((((((......((.(....).))........))))))).((((.(((.....((((((....)))))).((.(((....))).)).....))).))))... ( -21.04) >DroSim_CAF1 26450 103 - 1 UAGAGUAUCCAUUGGCCUAUUGACCUUUUACGUUAUUCUAUUCCUACCAUUUUCGUUUCCUAUUGGGAAUUGUUGGCAAUUGCUUACCCUUCUGGCAGGAUCC (((((((......((.(....).))........))))))).((((.(((.....((((((....)))))).((.(((....))).)).....))).))))... ( -21.04) >DroEre_CAF1 28227 101 - 1 UAGGGUAUCCAUUGGCCUAUUGCCCUUUUACGUUAUUUUAUUCCUACCAUUUUCGUUUCCUAUUGGGAAUUGUUGGCAAUUGCUUACCCUUC--GCAGGAUCC .((((((......(((..((((((.......((............)).......((((((....))))))....)))))).)))))))))..--......... ( -19.10) >DroYak_CAF1 35857 103 - 1 UAGGGUAUCCAUUGGCCUAUUGACCUUUUGCGUUAUCCUAUUCCUACCAUUUUCGUUUCCUAUUGGGAAUUGUUGGCAAUUGCUUACCCUUCUGGCAGGAUCC (((((((.(((..((.(....).))...)).).))))))).((((.(((.....((((((....)))))).((.(((....))).)).....))).))))... ( -25.00) >consensus UAGAGUAUCCAUUGGCCUAUUGACCUUUUACGUUAUUCUAUUCCUACCAUUUUCGUUUCCUAUUGGGAAUUGUUGGCAAUUGCUUACCCUUCUGGCAGGAUCC (((((((......((.(....).))........))))))).((((.(((.....((((((....)))))).((.(((....))).)).....))).))))... (-19.64 = -19.48 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:20 2006