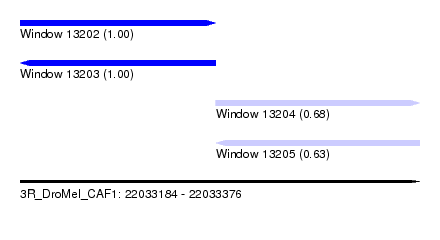
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,033,184 – 22,033,376 |
| Length | 192 |
| Max. P | 0.999992 |

| Location | 22,033,184 – 22,033,278 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 94.94 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -29.66 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.94 |
| SVM decision value | 5.20 |
| SVM RNA-class probability | 0.999978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
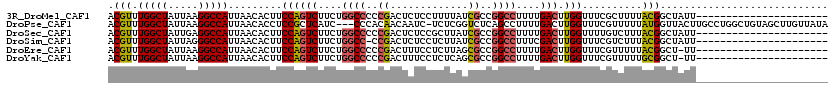
>3R_DroMel_CAF1 22033184 94 + 27905053 AUCGAUGAAGGGCUGUUUUUUUUU---UUCAGCUCUUCGAGCUGAUUUAUUUGGCCGGCACACACUGUGCGGCUUGUUUGUCCACUUGACGCUCAUC ......(((((((((.........---..)))))))))((((..........(((((.(((.....)))))))).....(((.....)))))))... ( -32.60) >DroSec_CAF1 31487 93 + 1 AUCGAUGAGGGGCUGUUUUU-UUU---UUCAGCUCUUCGAGCUGAUUUAUUUGGCCGGCACACACUGUGCGGCUUGUUUGUCCACUUGACGCUCAUC ...((((((((((((.....-...---..)))))).(((((..(((..((..(((((.(((.....)))))))).))..)))..)))))..)))))) ( -33.60) >DroSim_CAF1 23701 92 + 1 AUCGAUGAGGGGCUGUUUUU--UU---UUCAGCUCUUCGAGCUGAUUUAUUUGGCCGGCACACACUGUGCGGCUUGUUUGUCCACUUGACGCUCAUC ...((((((((((((.....--..---..)))))).(((((..(((..((..(((((.(((.....)))))))).))..)))..)))))..)))))) ( -33.70) >DroEre_CAF1 25438 89 + 1 AUCGAUGAGGGGAUGUUUUU--------UCAGCUCUUCGAGCUGAUUUAUUUGGCCGGCACACACUGUGCGGCUUGUUUGUCCACUUGACGCUCAUC ...((((((.(.........--------(((((((...))))))).......(((((.(((.....))))))))...............).)))))) ( -29.50) >DroYak_CAF1 33041 97 + 1 AUCGAUGAGGGGCUGUUUUUUUUUUUGUUCACCUCUUCGAGCUGAUUUAUUUGGCCGGCACACACUGUGCGGCUUGUUUGUCCACUUGACGCUCAUC ...((((((((((.............))))......(((((..(((..((..(((((.(((.....)))))))).))..)))..)))))..)))))) ( -27.62) >consensus AUCGAUGAGGGGCUGUUUUU_UUU___UUCAGCUCUUCGAGCUGAUUUAUUUGGCCGGCACACACUGUGCGGCUUGUUUGUCCACUUGACGCUCAUC ......(((((((((..............)))))))))((((..........(((((.(((.....)))))))).....(((.....)))))))... (-29.66 = -29.90 + 0.24)


| Location | 22,033,184 – 22,033,278 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 94.94 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -27.78 |
| Energy contribution | -27.82 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.97 |
| SVM decision value | 5.67 |
| SVM RNA-class probability | 0.999992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22033184 94 - 27905053 GAUGAGCGUCAAGUGGACAAACAAGCCGCACAGUGUGUGCCGGCCAAAUAAAUCAGCUCGAAGAGCUGAA---AAAAAAAAACAGCCCUUCAUCGAU ((((((((((.....)))......(((((((.....))).))))...........))))((((.((((..---.........)))).)))))))... ( -29.80) >DroSec_CAF1 31487 93 - 1 GAUGAGCGUCAAGUGGACAAACAAGCCGCACAGUGUGUGCCGGCCAAAUAAAUCAGCUCGAAGAGCUGAA---AAA-AAAAACAGCCCCUCAUCGAU ((((((.(((.....)))......(((((((.....))).))))........(((((((...))))))).---...-...........))))))... ( -29.50) >DroSim_CAF1 23701 92 - 1 GAUGAGCGUCAAGUGGACAAACAAGCCGCACAGUGUGUGCCGGCCAAAUAAAUCAGCUCGAAGAGCUGAA---AA--AAAAACAGCCCCUCAUCGAU ((((((.(((.....)))......(((((((.....))).))))........(((((((...))))))).---..--...........))))))... ( -29.50) >DroEre_CAF1 25438 89 - 1 GAUGAGCGUCAAGUGGACAAACAAGCCGCACAGUGUGUGCCGGCCAAAUAAAUCAGCUCGAAGAGCUGA--------AAAAACAUCCCCUCAUCGAU ((((((.(((.....)))......(((((((.....))).))))........(((((((...)))))))--------...........))))))... ( -29.50) >DroYak_CAF1 33041 97 - 1 GAUGAGCGUCAAGUGGACAAACAAGCCGCACAGUGUGUGCCGGCCAAAUAAAUCAGCUCGAAGAGGUGAACAAAAAAAAAAACAGCCCCUCAUCGAU ((((((.(((.....)))......(((((((.....))).))))........(((.(((...))).)))...................))))))... ( -25.00) >consensus GAUGAGCGUCAAGUGGACAAACAAGCCGCACAGUGUGUGCCGGCCAAAUAAAUCAGCUCGAAGAGCUGAA___AAA_AAAAACAGCCCCUCAUCGAU ((((((.(((.....)))......(((((((.....))).))))........(((((((...)))))))...................))))))... (-27.78 = -27.82 + 0.04)


| Location | 22,033,278 – 22,033,376 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.38 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -17.18 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
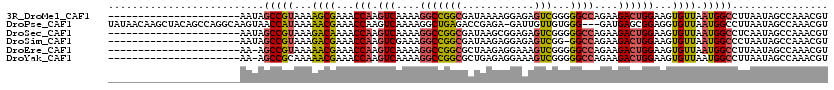
>3R_DroMel_CAF1 22033278 98 + 27905053 ACGUUUGGCUAUUAAGGCCAUUAACACUUCCAGUCUUCUGGCCCCCGACUCUCCUUUUAUCGCCGGCCUUUUGACUUGGUUUCGCUUUUACGGCUAUU---------------------- ..((((((((.....)))))..)))....((((((....((((..(((...........)))..))))....))).)))....(((.....)))....---------------------- ( -23.70) >DroPse_CAF1 96163 116 + 1 ACGUUUGGCUAUUAAGGCCAUUAACACCUCCGCUCAUC---CCCACAACAAUC-UCUCGGUCUCAGCCUUUUGACUUGGUUUCGUUUUUAUGGUUACUUGCCUGGCUGUAGCUUGUUAUA ..((((((((.....)))))..))).............---.....(((((.(-(..(((((..((((.........))))..........(((.....))).))))).)).)))))... ( -20.80) >DroSec_CAF1 31580 98 + 1 ACGUUUGGCUAUUGAGGCCAUUAACACUUCCAGUCUUCUGGCCCCCGACUCUCCGCUUAUCGCCGGCCUUUUGACUUGGUUUUGUCUUUACGGCUAUU---------------------- .(((.(((((.....)))))...(((...((((((....((((..(((...........)))..))))....))).)))...)))....)))......---------------------- ( -24.30) >DroSim_CAF1 23793 97 + 1 ACGUUUGGCUAUUAGGGCCAUUAACACUUCCAGUCUUCUGGCC-CCGACUCUCCUCUUAUCGCCGGCCUUUCGACUUGGUUUCGUCUUUACGGCUAUU---------------------- ..((((((((.....)))))..)))....((((((....((((-.(((...........)))..))))....))).)))....(((.....)))....---------------------- ( -21.70) >DroEre_CAF1 25527 97 + 1 ACGUUUGGCUAUUAAGGCCAUUAACACUUCCAGUCUUCUGGCCCCCGACUUUCCUCUUAGCGCCGGCCUUUUGACUUGGUUUCGUUUUUACGGCU-UU---------------------- ..((((((((.....)))))..)))....((((((....((((..((.((........))))..))))....))).)))..((((....))))..-..---------------------- ( -24.70) >DroYak_CAF1 33138 97 + 1 ACGUUUGGCUAUUAAGGCCAUUAACACUUCCAGUCUUCUGGCCCCCGACUUUCCUCUCAGCGCCGGCCUUUUGACUUGGUUUCGUUUUUGCGGCU-UU---------------------- ..((((((((.....)))))..)))....((((((....((((..((.((........))))..))))....))).)))..((((....))))..-..---------------------- ( -24.30) >consensus ACGUUUGGCUAUUAAGGCCAUUAACACUUCCAGUCUUCUGGCCCCCGACUCUCCUCUUAGCGCCGGCCUUUUGACUUGGUUUCGUUUUUACGGCUAUU______________________ .(((.(((((.....))))).........((((((....((((..((.............))..))))....))).)))..........)))............................ (-17.18 = -17.32 + 0.14)
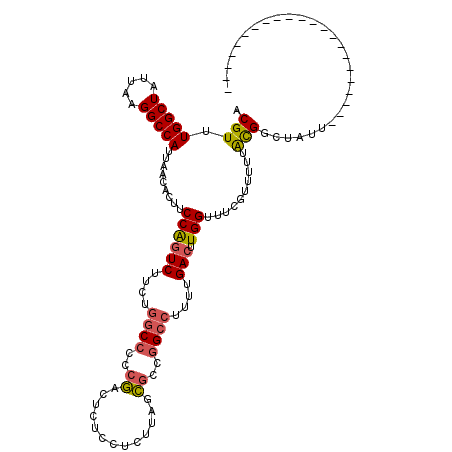
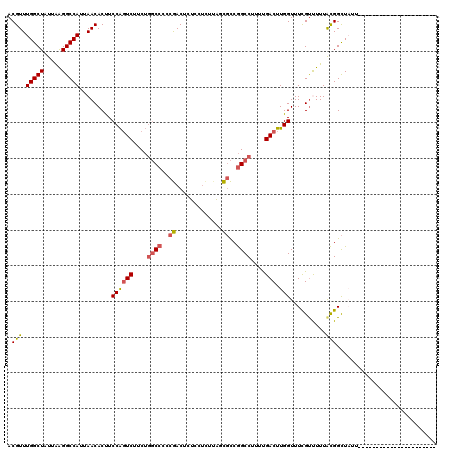
| Location | 22,033,278 – 22,033,376 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.38 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -17.39 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22033278 98 - 27905053 ----------------------AAUAGCCGUAAAAGCGAAACCAAGUCAAAAGGCCGGCGAUAAAAGGAGAGUCGGGGGCCAGAAGACUGGAAGUGUUAAUGGCCUUAAUAGCCAAACGU ----------------------....(((((...((((...(((.(((....((((..((((.........))))..))))....))))))...)))).)))))................ ( -24.80) >DroPse_CAF1 96163 116 - 1 UAUAACAAGCUACAGCCAGGCAAGUAACCAUAAAAACGAAACCAAGUCAAAAGGCUGAGACCGAGA-GAUUGUUGUGGG---GAUGAGCGGAGGUGUUAAUGGCCUUAAUAGCCAAACGU ((((((((.((.(((((.((.......))........((.......))....)))))))..(....-).)))))))).(---..((.((.(((((.......)))))....))))..).. ( -23.60) >DroSec_CAF1 31580 98 - 1 ----------------------AAUAGCCGUAAAGACAAAACCAAGUCAAAAGGCCGGCGAUAAGCGGAGAGUCGGGGGCCAGAAGACUGGAAGUGUUAAUGGCCUCAAUAGCCAAACGU ----------------------....(((.....(((........)))....))).(((......(((....)))(((((((...(((.......)))..)))))))....)))...... ( -26.90) >DroSim_CAF1 23793 97 - 1 ----------------------AAUAGCCGUAAAGACGAAACCAAGUCGAAAGGCCGGCGAUAAGAGGAGAGUCGG-GGCCAGAAGACUGGAAGUGUUAAUGGCCCUAAUAGCCAAACGU ----------------------....(((.....(((........)))....))).((((((.........)))((-(((((...(((.......)))..)))))))....)))...... ( -29.10) >DroEre_CAF1 25527 97 - 1 ----------------------AA-AGCCGUAAAAACGAAACCAAGUCAAAAGGCCGGCGCUAAGAGGAAAGUCGGGGGCCAGAAGACUGGAAGUGUUAAUGGCCUUAAUAGCCAAACGU ----------------------..-.(((((...((((...(((.(((....((((..((((........)).))..))))....))))))...)))).)))))................ ( -23.50) >DroYak_CAF1 33138 97 - 1 ----------------------AA-AGCCGCAAAAACGAAACCAAGUCAAAAGGCCGGCGCUGAGAGGAAAGUCGGGGGCCAGAAGACUGGAAGUGUUAAUGGCCUUAAUAGCCAAACGU ----------------------..-...........................(((((((.((....))...))))(((((((...(((.......)))..)))))))....)))...... ( -24.60) >consensus ______________________AAUAGCCGUAAAAACGAAACCAAGUCAAAAGGCCGGCGAUAAGAGGAGAGUCGGGGGCCAGAAGACUGGAAGUGUUAAUGGCCUUAAUAGCCAAACGU ..........................(((((...((((...(((.(((....(((((((............)))...))))....))))))...)))).)))))................ (-17.39 = -17.50 + 0.11)
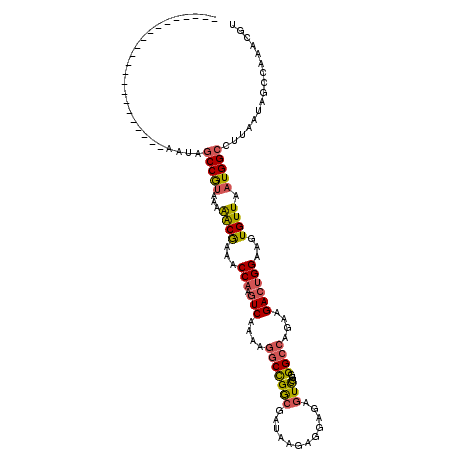
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:16 2006