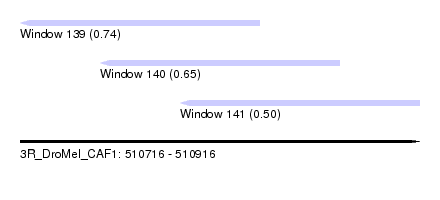
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 510,716 – 510,916 |
| Length | 200 |
| Max. P | 0.739464 |

| Location | 510,716 – 510,836 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -23.67 |
| Energy contribution | -23.79 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 510716 120 - 27905053 AUGCAAAAUACUUUUGCUCUCAUACCUUGAGAGAGCCAACGAAGCAUUCAAUUCUAGCUUGGCCGCGAUCCCCUUUAUUCACACGCACUACGCCCCUAAAGGGGGAAGCUAGGCAAAAAU ..(((((.....)))))(((((.....)))))..(((..(.((((...........)))).)..((..(((((((((.......((.....))...)))))))))..))..)))...... ( -31.80) >DroSim_CAF1 37516 120 - 1 AUGCAAAAUACCUUUGCUCUCAUACCUUGAGAGACCCAACGAAGCAUUCAAUUAUAGCUUGGCCGCGAUCCCCUUCAUUCACACGCACUACGCCCCUAAAAGGGGGAACUUGGCAACAAU ..(((((.....)))))(((((.....))))).........((((...........)))).((((.(.(((((((.........((.....)).......))))))).).))))...... ( -27.39) >DroEre_CAF1 41743 120 - 1 ACUCAAAAUACUUUUGCUCUCAUACCUUGAGAGACCCAACGAAGCAUUUAAUUCUAGCUGGGUCGCUAUCCCCUUUAUUUACAGGCACUACGCCCCUAAAAGGGGAAACUUUGCAACAAU .............(((((((((.....)))))((((((..(((........)))....))))))....((((((((...((..(((.....)))..))))))))))......)))).... ( -32.90) >consensus AUGCAAAAUACUUUUGCUCUCAUACCUUGAGAGACCCAACGAAGCAUUCAAUUCUAGCUUGGCCGCGAUCCCCUUUAUUCACACGCACUACGCCCCUAAAAGGGGAAACUUGGCAACAAU ................((((((.....)))))).......................(((((((.(((................)))......((((.....))))..)))))))...... (-23.67 = -23.79 + 0.12)

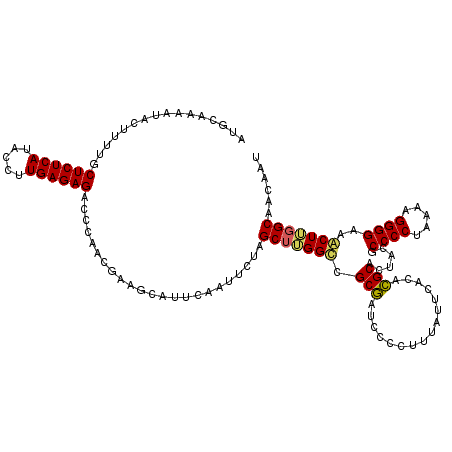
| Location | 510,756 – 510,876 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.64 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -18.52 |
| Energy contribution | -18.97 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 510756 120 - 27905053 UUCUCACUCCCCACGUCCGCUGCCCGCCCCCACAGUUGGCAUGCAAAAUACUUUUGCUCUCAUACCUUGAGAGAGCCAACGAAGCAUUCAAUUCUAGCUUGGCCGCGAUCCCCUUUAUUC .................(((.(((..........((((((..(((((.....)))))(((((.....)))))..)))))).((((...........))))))).)))............. ( -30.80) >DroSim_CAF1 37556 116 - 1 UGCUCACUCCCCACGUCCGCUGC----CCCCACAGUUGGCAUGCAAAAUACCUUUGCUCUCAUACCUUGAGAGACCCAACGAAGCAUUCAAUUAUAGCUUGGCCGCGAUCCCCUUCAUUC .................(((.((----(......(((((...(((((.....)))))(((((.....)))))...))))).((((...........))))))).)))............. ( -26.40) >DroEre_CAF1 41783 116 - 1 UUCUUACUCCCCAAGCGCGCAGC----CCCCACAGUUGGAACUCAAAAUACUUUUGCUCUCAUACCUUGAGAGACCCAACGAAGCAUUUAAUUCUAGCUGGGUCGCUAUCCCCUUUAUUU .............(((((.((((----.......(((((....((((.....))))((((((.....))))))..)))))(((........)))..)))).).))))............. ( -28.20) >consensus UUCUCACUCCCCACGUCCGCUGC____CCCCACAGUUGGCAUGCAAAAUACUUUUGCUCUCAUACCUUGAGAGACCCAACGAAGCAUUCAAUUCUAGCUUGGCCGCGAUCCCCUUUAUUC .................(((.((...........(((((....((((.....))))((((((.....))))))..))))).((((...........)))).)).)))............. (-18.52 = -18.97 + 0.45)

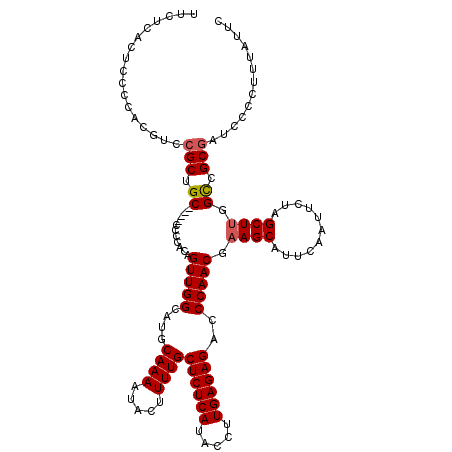
| Location | 510,796 – 510,916 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -17.33 |
| Energy contribution | -17.33 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
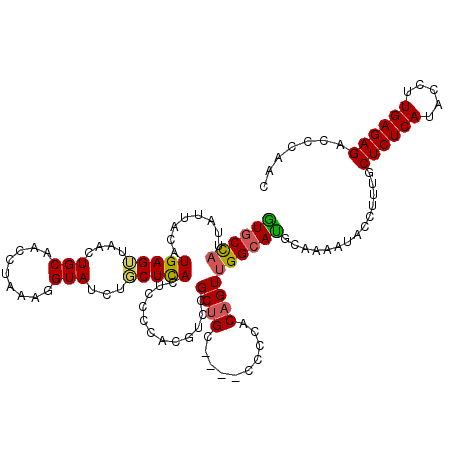
>3R_DroMel_CAF1 510796 120 - 27905053 AUGCCAUUAUUACAUGAGAUAACUGCAACCUAAAGGUAUCUUCUCACUCCCCACGUCCGCUGCCCGCCCCCACAGUUGGCAUGCAAAAUACUUUUGCUCUCAUACCUUGAGAGAGCCAAC .(((..(((((......)))))..))).......(((.....................((((((.((.......)).)))).))............((((((.....)))))).)))... ( -24.50) >DroSec_CAF1 37516 116 - 1 GUGCCAUUAUUACUUGAGUUAACUGCAACCUAAAGGUAUCUGCUCACUCCCCACGUCCGCUGC----CCCCACAGUUGGCAUGCAAAAUACCUUUGCUCUCAUACCUUGAGAGACCCAAC ((((((.........((((.....((((((....)))...)))..)))).........((((.----.....))))))))))(((((.....)))))(((((.....)))))........ ( -25.50) >DroSim_CAF1 37596 116 - 1 GUGCCAUUAUUACAUGAGUUAACUGCAACCUAAAGGUAUCUGCUCACUCCCCACGUCCGCUGC----CCCCACAGUUGGCAUGCAAAAUACCUUUGCUCUCAUACCUUGAGAGACCCAAC ((((((.........((((.....((((((....)))...)))..)))).........((((.----.....))))))))))(((((.....)))))(((((.....)))))........ ( -25.50) >DroEre_CAF1 41823 114 - 1 GUGCUUUUAUUACAUGAG-UAACUGCAACCUAAA-GUAUCUUCUUACUCCCCAAGCGCGCAGC----CCCCACAGUUGGAACUCAAAAUACUUUUGCUCUCAUACCUUGAGAGACCCAAC ((((((...((((....)-)))...........(-(((......))))....)))))).....----.......(((((....((((.....))))((((((.....))))))..))))) ( -21.60) >consensus GUGCCAUUAUUACAUGAGUUAACUGCAACCUAAAGGUAUCUGCUCACUCCCCACGUCCGCUGC____CCCCACAGUUGGCAUGCAAAAUACCUUUGCUCUCAUACCUUGAGAGACCCAAC ((((((........(((((....(((.........)))...)))))............((((..........))))))))))..............((((((.....))))))....... (-17.33 = -17.33 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:35 2006