
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,023,844 – 22,024,015 |
| Length | 171 |
| Max. P | 0.914247 |

| Location | 22,023,844 – 22,023,935 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -18.30 |
| Energy contribution | -18.14 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
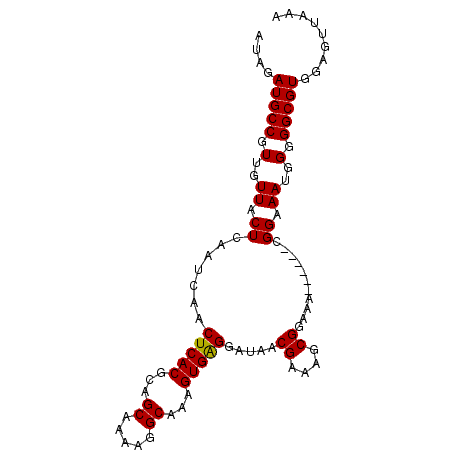
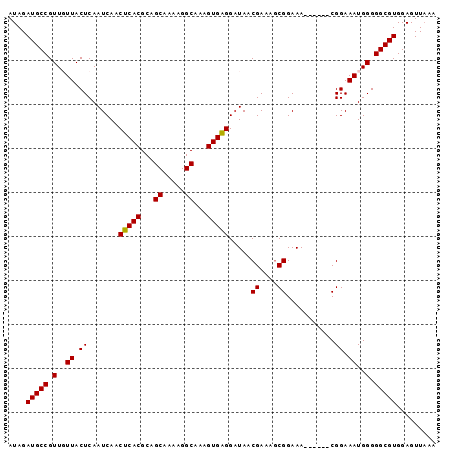
>3R_DroMel_CAF1 22023844 91 + 27905053 AUAGAUGCCGUUGUUACUCAAUCAACUCACGCAGCAAAAGGCAAAGUGAGGAUAACGAAAGCGGAAA------CGGAAAUGGGGGCGUGGAGUUAAA ....(((((.(..((..........(((((...((.....))...))))).....(....)((....------))..))..).)))))......... ( -23.10) >DroSec_CAF1 22286 91 + 1 AUAGAUGCCGUUGUUACUCAAUCAACUCACGCAGCAAAAGGCAAAGUGGGGAUAACGAAAGCGGAAA------CGGAAAUGGGGGCGUGGAGUUAAA ....(((((.(..((..........(((((...((.....))...))))).....(....)((....------))..))..).)))))......... ( -22.20) >DroSim_CAF1 14347 91 + 1 AUAGAUGCCGUUGUUACUCAAUCAACUCACGCAGCAAAAGGCAAAGUGAGGAUAACGAAAGCGGAAA------CGGAAAUGGGGGCGUGGAGUUAAA ....(((((.(..((..........(((((...((.....))...))))).....(....)((....------))..))..).)))))......... ( -23.10) >DroEre_CAF1 16125 91 + 1 AUAGAUGCCGUUCUUACUCAAUCAACUCACGCAGCAAAAGGCACAGUGAGGAUAACGAAUGCGGAAA------CGGAAAUGGGGGCGUGGAGUUAAA .(((.(.(((((((((.........(((((...((.....))...))))).........(.((....------)).)..)))))))).).).))).. ( -20.20) >DroYak_CAF1 23566 97 + 1 AUAGAUGCCGUUCUUACUCAAUCAACUCACGCAGCGAAAGGCAAAGUGAGGCUAACGAAUACGGAAACGGAAACGGAAAUGGGGGCGUGGAGUUUAA .(((((.(((((((((.((......(((((...((.....))...))))).....((....((....))....))))..)))))))).)..))))). ( -23.60) >consensus AUAGAUGCCGUUGUUACUCAAUCAACUCACGCAGCAAAAGGCAAAGUGAGGAUAACGAAAGCGGAAA______CGGAAAUGGGGGCGUGGAGUUAAA ....(((((.(..((.((.......(((((...((.....))...))))).....((....))...........)).))..).)))))......... (-18.30 = -18.14 + -0.16)

| Location | 22,023,905 – 22,024,015 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 95.91 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -23.20 |
| Energy contribution | -23.44 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
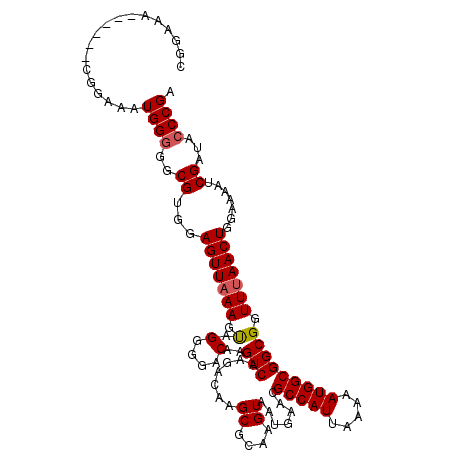
>3R_DroMel_CAF1 22023905 110 + 27905053 CGGAAA------CGGAAAUGGGGGCGUGGAGUUAAAGUAGCGGACAAGUCAAGACAAGCGCAAGUAAUGAACGCCAUUAAAAAUGGCGGCGGUUUAACUGGAAAAUCGAUAACCGA ((....------))....(((..(((...(((((((...((................((....))......((((((.....))))))))..))))))).......)).)..))). ( -24.60) >DroSec_CAF1 22347 110 + 1 CGGAAA------CGGAAAUGGGGGCGUGGAGUUAAAGUAGGGGACAAGUCAAGACAAGCGCAAGUAAUGAACGCCAUUAAAAAUGGCGGCGGUUUAACUGGAAAAUCGAUACCCGA ((....------))....((((..((...(((((((.(.(....)..(((.......((....)).......(((((.....))))))))).))))))).......))...)))). ( -28.70) >DroSim_CAF1 14408 110 + 1 CGGAAA------CGGAAAUGGGGGCGUGGAGUUAAAGUAGGGGACAAGUCAAGACAAGCGCAAGUAAUGAACGCCAUUAAAAAUGGCGGCGGUUUAACUGGAAAAUCGAUACCCGA ((....------))....((((..((...(((((((.(.(....)..(((.......((....)).......(((((.....))))))))).))))))).......))...)))). ( -28.70) >DroEre_CAF1 16186 110 + 1 CGGAAA------CGGAAAUGGGGGCGUGGAGUUAAAGCAGGGGACAAGUCAAGACAAGCGCAAGUAAUGAAAGCCAUUAAAAAUGGCGGCGGUUUAACUGGAAAAUCGAUACCCGA ((....------))....((((..((...(((((((.(.(....)..(((.......((....)).......(((((.....))))))))).))))))).......))...)))). ( -30.70) >DroYak_CAF1 23627 116 + 1 CGGAAACGGAAACGGAAAUGGGGGCGUGGAGUUUAAGUAGGGGACAAGUCAAGACAAGCGCAAGUAAUGAAAGCCAUUAAAAAUGGCGGCGGUUUAACUGGAAAAUCGAUACCCGA ((....((....))....)).(((..(.((.(((.(((.(....)..(((.......((....)).......(((((.....))))))))......)))..))).)).)..))).. ( -28.80) >consensus CGGAAA______CGGAAAUGGGGGCGUGGAGUUAAAGUAGGGGACAAGUCAAGACAAGCGCAAGUAAUGAACGCCAUUAAAAAUGGCGGCGGUUUAACUGGAAAAUCGAUACCCGA ..................((((..((...(((((((.(.(....)..(((.......((....)).......(((((.....))))))))).))))))).......))...)))). (-23.20 = -23.44 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:02 2006