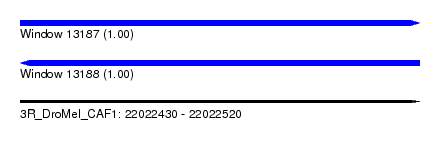
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 22,022,430 – 22,022,520 |
| Length | 90 |
| Max. P | 0.999867 |

| Location | 22,022,430 – 22,022,520 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -26.36 |
| Consensus MFE | -23.26 |
| Energy contribution | -23.46 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.31 |
| SVM RNA-class probability | 0.999867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22022430 90 + 27905053 UUU-GG------UAUGUGUGCGUGUGUGUUUAAUUACACAUUACGUAUACGCCACAUUUGACGCAUUGUGGCUGCUGUACUGUGGCUUGUAUAUUUU ...-((------((.((((((((((((((......))))).)))))))))((((((..........))))))))))((((........))))..... ( -29.50) >DroSec_CAF1 20669 89 + 1 UUUGGG------UAUGUGUGCUUG--UGUUUAAUUACACAUUACGUAUACGCCACAUUUGACGCAUUGUGGCUGUUGUACUGUGGCUUGUAUAUUUU .....(------((((((((..((--(((......))))).)))))))))((((((..((((((......)).))))...))))))........... ( -24.60) >DroSim_CAF1 12726 89 + 1 UUUUGG------UAUGUGUGCGUG--UGUUUAAUUACACAUUACGUAUACGCCACAUUUGACGCAUUGUGGCUGCUGUACUGUGGCUUGUAUAUUUU ....((------((.(((((((((--(((......))))...))))))))((((((..........))))))))))((((........))))..... ( -25.10) >DroEre_CAF1 14508 88 + 1 GU-ACG------UUUGUGUGCGUG--UGUUUAAUUACACAUUACGUAUACGCCACAUUUGACGCAUUGUGGCUACUGUACUGUGGCUUGUAUAUUUU ((-((.------...(((((((((--(((......))))...))))))))((((((..........))))))....))))................. ( -25.80) >DroYak_CAF1 21906 94 + 1 GU-ACUUACAUAUGUGUGUGCGUG--UGUUUAAUUACACAUUACGUAUACGCCACAUUUGACGCAUUGUGGCUGCUGUACUGUGGCUUGUAUAUUUU ((-((...((((.(((((((((((--(((......))))...))))))))((((((..........))))))......))))))....))))..... ( -26.80) >consensus UUUAGG______UAUGUGUGCGUG__UGUUUAAUUACACAUUACGUAUACGCCACAUUUGACGCAUUGUGGCUGCUGUACUGUGGCUUGUAUAUUUU ...............(((((((((..(((......)))...)))))))))((((((..........))))))...(((((........))))).... (-23.26 = -23.46 + 0.20)

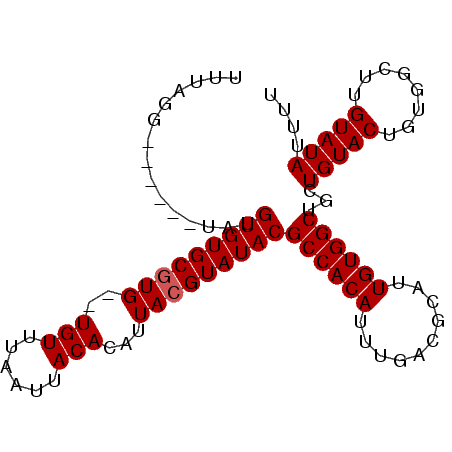
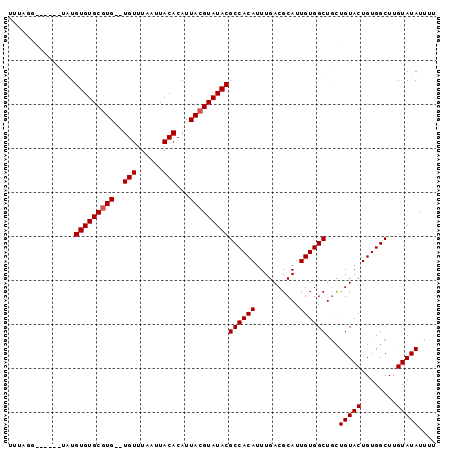
| Location | 22,022,430 – 22,022,520 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -20.13 |
| Consensus MFE | -16.98 |
| Energy contribution | -17.02 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.87 |
| SVM RNA-class probability | 0.999674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 22022430 90 - 27905053 AAAAUAUACAAGCCACAGUACAGCAGCCACAAUGCGUCAAAUGUGGCGUAUACGUAAUGUGUAAUUAAACACACACGCACACAUA------CC-AAA ....(((((..((((((.....(((.......)))......)))))))))))(((..(((((......))))).)))........------..-... ( -21.00) >DroSec_CAF1 20669 89 - 1 AAAAUAUACAAGCCACAGUACAACAGCCACAAUGCGUCAAAUGUGGCGUAUACGUAAUGUGUAAUUAAACA--CAAGCACACAUA------CCCAAA ....(((((..((((((........((......))......))))))))))).(((.(((((..((.....--.))..)))))))------)..... ( -17.74) >DroSim_CAF1 12726 89 - 1 AAAAUAUACAAGCCACAGUACAGCAGCCACAAUGCGUCAAAUGUGGCGUAUACGUAAUGUGUAAUUAAACA--CACGCACACAUA------CCAAAA ....(((((..((((((.....(((.......)))......))))))))))).((..(((((......)))--)).)).......------...... ( -20.00) >DroEre_CAF1 14508 88 - 1 AAAAUAUACAAGCCACAGUACAGUAGCCACAAUGCGUCAAAUGUGGCGUAUACGUAAUGUGUAAUUAAACA--CACGCACACAAA------CGU-AC ...........((((((.....(((.......)))......))))))...(((((..(((((.........--...)))))...)------)))-). ( -19.30) >DroYak_CAF1 21906 94 - 1 AAAAUAUACAAGCCACAGUACAGCAGCCACAAUGCGUCAAAUGUGGCGUAUACGUAAUGUGUAAUUAAACA--CACGCACACACAUAUGUAAGU-AC ......(((..((((((.....(((.......)))......))))))...((((((.(((((.........--.......))))))))))).))-). ( -22.59) >consensus AAAAUAUACAAGCCACAGUACAGCAGCCACAAUGCGUCAAAUGUGGCGUAUACGUAAUGUGUAAUUAAACA__CACGCACACAUA______CCUAAA ....(((((..((((((.....(((.......)))......))))))))))).....(((((..............)))))................ (-16.98 = -17.02 + 0.04)

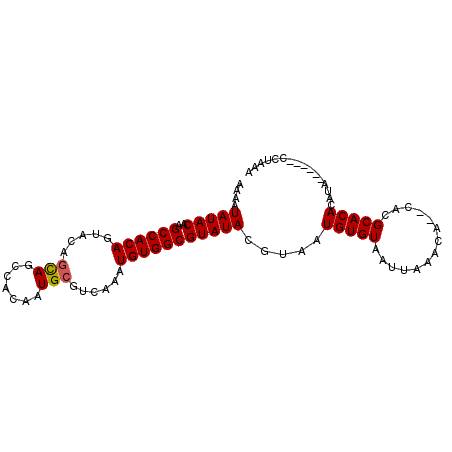
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:05:00 2006