| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,805,113 – 2,805,304 |
| Length | 191 |
| Max. P | 0.683554 |

| Location | 2,805,113 – 2,805,212 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 84.44 |
| Mean single sequence MFE | -24.69 |
| Consensus MFE | -17.10 |
| Energy contribution | -17.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
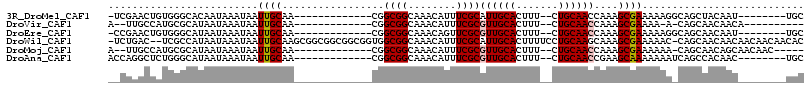
>3R_DroMel_CAF1 2805113 99 - 27905053 GCGGGACAGCACCCGCUUCCACUUCCCUCCCACAUUUUCGUCUGCCUGGGUCCUGCCCAC---------UUUGCCCCCGCAAUUCCCACCUCGGUGACC--------CUCCCCACU (((((......))))).......................(((.(((((((.....)))).---------.((((....))))..........)))))).--------......... ( -22.70) >DroSec_CAF1 80533 98 - 1 GCGGGACAGCACCCGCUUCCACUUCCCUGCCACAUUUUCGUCUGCC-GGGUCCUGCCCAC---------UUUGCCCCCGCAUUUCCCACCUCGGCGACC--------CUCUGCACU (((((......))))).......................(((.(((-(((.....)))..---------..(((....)))...........)))))).--------......... ( -24.00) >DroSim_CAF1 84708 98 - 1 GCGGGACAGCACCCGCUUACACUUCCCUCCCACAUUUUCGUCUGCC-GGGUCCUGCCCAC---------UUUGCCCCCGCAUUUCCCACCUCGGCGACC--------CGCCCCACU (((((.(((.((((((..((...................))..).)-)))).))).....---------.(((((.................)))))))--------)))...... ( -24.04) >DroEre_CAF1 82324 106 - 1 GCGGGACAGCACCCGCUUCCACUUCCCUCCCACAUUUUCGUCUGCC-GGGUCCUGCCCACUUUGCCCACUUUGCCCCCGCAUUUCCCACCUCA---------GCGAGCCCCGCACU (((((..(((....)))...................((((.(((..-(((.....))).............(((....)))..........))---------))))).)))))... ( -23.80) >DroYak_CAF1 83609 106 - 1 GCGGGACAGCACCCGCUUGCACUUCCCUCCCACAUUUUCGUACGCC-GGGUCCUGCCCAC---------UUUGCCCCCGCAUUUCCCACCUCCGCAACCUCCGCGGUCCCCGCGCU (((((.(((.((((((.(((...................))).).)-)))).))))))..---------..(((....)))............))......(((((...))))).. ( -28.91) >consensus GCGGGACAGCACCCGCUUCCACUUCCCUCCCACAUUUUCGUCUGCC_GGGUCCUGCCCAC_________UUUGCCCCCGCAUUUCCCACCUCGGCGACC________CCCCGCACU (((((..(((....)))..............................(((.....))).................))))).................................... (-17.10 = -17.10 + -0.00)

| Location | 2,805,212 – 2,805,304 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.80 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -12.29 |
| Energy contribution | -12.48 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2805212 92 - 27905053 -UCGAACUGUGGGCACAAUAAAUAAUUGCAA-------------CGGCGGCAAACAUUUCGCAUUGCACUUU--CUGCAACCAAAGCGAAAAAGGCAGCUACAAU--------UGC -......(((((((...........((((..-------------.....))))...((((((.(((((....--.))))).....))))))...))..)))))..--------... ( -17.00) >DroVir_CAF1 134382 87 - 1 A--UUGCCAUGCGCAUAAUAAAUAAUUGCAA-------------CGGCGGCAAACAUUUCGCGUUGCACUUU--CUGCAACCAAAGCGAAAA-A-CAGCAACAACA---------- .--(((((.((.(((...........)))..-------------))..)))))...((((((((((((....--.))))))....)))))).-.-...........---------- ( -20.30) >DroEre_CAF1 82430 92 - 1 -CCGAACUGUGGGCAUAAUAAAUAAUUGCAA-------------CGGCGGCAAACAGUUCGCGUUGCACUUU--CUGCAACCAAAGCGAAAAAGGCAGCAACAAU--------UGC -.((((((((..((..(((.....)))((..-------------..)).))..)))))))).(((((.((((--.(((.......)))..)))))))))......--------... ( -24.30) >DroWil_CAF1 102167 112 - 1 -UCUGAC--UCGCCAUAAUAAAUAAUUGCAAGCGGCGGCGGCGGUGGCGGCAAACAUUUCGCAUUGCACUUUUCCUGCAAGCAAAGCGAAAAAC-CAGCAACAACAACAACAACAC -......--................((((..((.((.((....)).)).)).....((((((.((((.............)))).))))))...-..))))............... ( -24.22) >DroMoj_CAF1 140148 93 - 1 A--UUGCCAUGCGCAUAAUAAAUAAUUGCAA-------------CGGCGGCAAACAUUUCGCGUUGCACUUU--CUGCAACCAAAGCGAAAAAA-CAGCAACAGCAACAAC----- .--(((((.((.(((...........)))..-------------))..)))))...((((((((((((....--.))))))....))))))...-..((....))......----- ( -21.50) >DroAna_CAF1 74321 93 - 1 ACCAGGCUCUGGGCAUAAUAAAUAAUUGCAA-------------CGGCGGCAAACAUUUCGCGUUGCACUUU--CUGCAACCGAAGCAAAAAAAUCAGCCACAAC--------UGC .((((...))))(((...........)))..-------------.((((.....).(((((.((((((....--.)))))))))))...........))).....--------... ( -21.50) >consensus __CUAACUAUGGGCAUAAUAAAUAAUUGCAA_____________CGGCGGCAAACAUUUCGCGUUGCACUUU__CUGCAACCAAAGCGAAAAAA_CAGCAACAAC________UGC .........................((((.................((((........))))((((((.......))))))....))))........................... (-12.29 = -12.48 + 0.20)
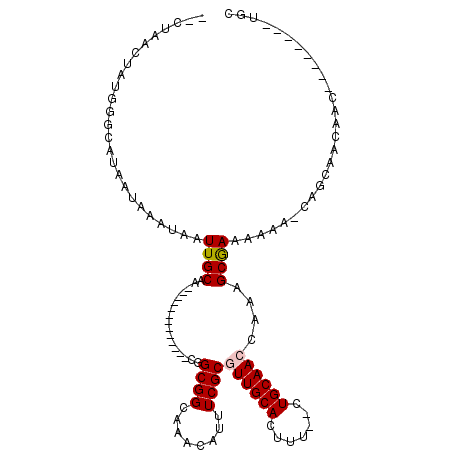
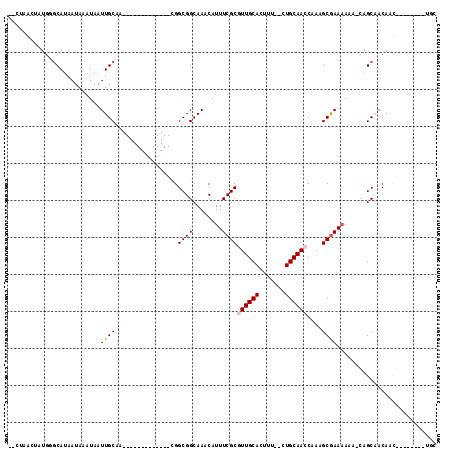
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:55 2006