
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,997,995 – 21,998,460 |
| Length | 465 |
| Max. P | 0.996133 |

| Location | 21,997,995 – 21,998,115 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.71 |
| Mean single sequence MFE | -37.55 |
| Consensus MFE | -28.41 |
| Energy contribution | -27.98 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21997995 120 + 27905053 GCGAGAUAAAAAAAAGUUUAAAACAAACUUGAGUUUUUCUCCCUGUGCAUAAAACCUGAAGGGGCACCAAAGCCCUUGCAAAUGUCGUGCGUUAGUAUGGCAGCAGGGCCAUUUAGUUGG ((((((.(((...((((((.....))))))...))).))))(((((..........((.((((((......)))))).))..((((((((....)))))))))))))))........... ( -37.40) >DroSim_CAF1 34320 119 + 1 GGGAGCUAAAAUAAAUUUUG-AAUAGACUUGAGUUUUUCUCCCUGUGCAUAAAACCUGAAGGGGCACCAAAGCCCUUGCAAAUGUCGUGCGUUAGUAUGGCAGCAGGGCCAUUUAGUUGG ...(((((((.........(-((.((((....))))))).((((((..........((.((((((......)))))).))..((((((((....))))))))))))))...))))))).. ( -36.20) >DroEre_CAF1 40056 118 + 1 GCGAGCUAA-ACGAGUUUUG-CAUAGACCUGAGUUUUUCUCCCUGUGCACGAAGCCUGAAGGGGCAGCAAAGCCCUUGCAAAUGUCGUGCGUUAGUACGGCGGCAUGGCCAUUUAGUUGG .(.((((((-(.(.(((.((-(((((....(((.....))).)))))))....(((((.((((((......)))))).))...(((((((....))))))))))..)))).))))))).) ( -41.60) >DroYak_CAF1 38686 113 + 1 -----CUAA-AUAAGUUUUG-GAUUGACUUGAGUUUUUCUCGCUGUGCAUGAAACCUGAAGGGGCACCAAAGCCCUUGCAAAUGUCGUGCGUUAGUAUGGCGGCAGGGCCAUUUAGUUGG -----((((-(((((((...-....)))))(((.....)))(((.(((........((.((((((......)))))).))..((((((((....))))))))))).))).)))))).... ( -35.00) >consensus GCGAGCUAA_AUAAAUUUUG_AAUAGACUUGAGUUUUUCUCCCUGUGCAUAAAACCUGAAGGGGCACCAAAGCCCUUGCAAAUGUCGUGCGUUAGUAUGGCAGCAGGGCCAUUUAGUUGG ..............................(((.....))).((.(((........((.((((((......)))))).))..((((((((....))))))))))).))(((......))) (-28.41 = -27.98 + -0.44)

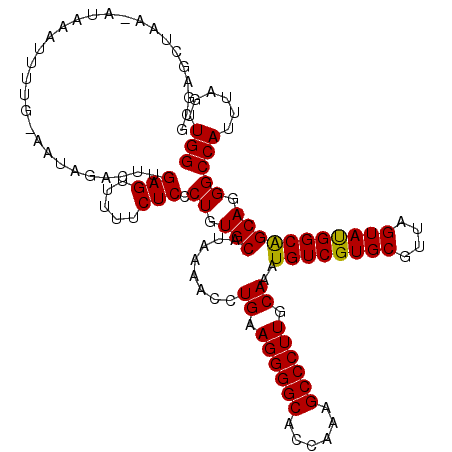
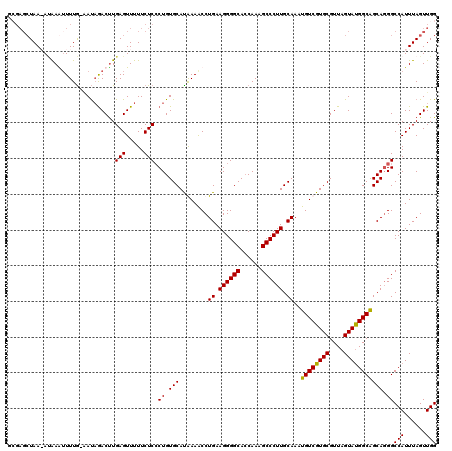
| Location | 21,998,035 – 21,998,155 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.42 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -31.46 |
| Energy contribution | -33.14 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21998035 120 + 27905053 CCCUGUGCAUAAAACCUGAAGGGGCACCAAAGCCCUUGCAAAUGUCGUGCGUUAGUAUGGCAGCAGGGCCAUUUAGUUGGGACCGGCGUCACAUUACGUAUACGCAAUGUGCACGAAAAU ((((((..........((.((((((......)))))).))..((((((((....))))))))))))))(((......)))......(((((((((.((....)).)))))).)))..... ( -42.50) >DroSim_CAF1 34359 120 + 1 CCCUGUGCAUAAAACCUGAAGGGGCACCAAAGCCCUUGCAAAUGUCGUGCGUUAGUAUGGCAGCAGGGCCAUUUAGUUGGGACCGGCGUCACAUUACGUAUACGCAAUGUGCACGAAAAU ((((((..........((.((((((......)))))).))..((((((((....))))))))))))))(((......)))......(((((((((.((....)).)))))).)))..... ( -42.50) >DroEre_CAF1 40094 120 + 1 CCCUGUGCACGAAGCCUGAAGGGGCAGCAAAGCCCUUGCAAAUGUCGUGCGUUAGUACGGCGGCAUGGCCAUUUAGUUGGGACCGGCGUCACAUUACGUAUACGCAAUGUGCACGAAAAU ...((((((((..(((((.((((((......)))))).))...(((((((....)))))))))).((((((......)))..)))((((.((.....))..))))..))))))))..... ( -44.60) >DroYak_CAF1 38719 120 + 1 CGCUGUGCAUGAAACCUGAAGGGGCACCAAAGCCCUUGCAAAUGUCGUGCGUUAGUAUGGCGGCAGGGCCAUUUAGUUGGGACCGGCGUCACAUUACGUAUACGCAAUGUGCACGAAAAU .((((..(......((((.((((((......)))))).....((((((((....)))))))).)))).(((......))))..))))((((((((.((....)).)))))).))...... ( -41.70) >DroAna_CAF1 44412 112 + 1 --------GUGAAAGUUGAUGCGCCACUGAAGCCAUCCCAAAUGUCGUGCGUUAGUGUUGCGGCAGAGCCAUUUAGUUGGGACCGGCGUCACAUUACGUAUACGCAGUGUGCACGAAAAU --------.......(((.(((((.((((..(((.((((((..((.(..(....)..).))(((...)))......))))))..))).........((....)))))))))))))).... ( -31.60) >consensus CCCUGUGCAUGAAACCUGAAGGGGCACCAAAGCCCUUGCAAAUGUCGUGCGUUAGUAUGGCGGCAGGGCCAUUUAGUUGGGACCGGCGUCACAUUACGUAUACGCAAUGUGCACGAAAAU ((((((..........((.((((((......)))))).))..((((((((....))))))))))))))(((......)))......(((((((((.((....)).)))))).)))..... (-31.46 = -33.14 + 1.68)

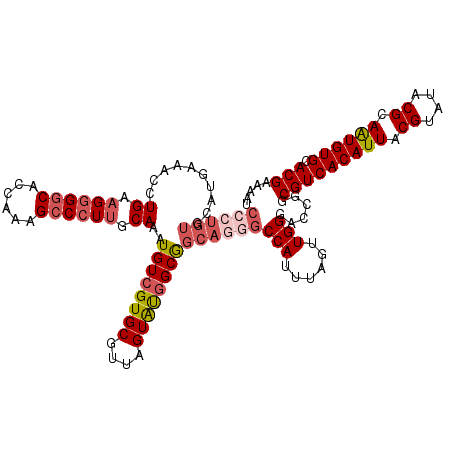
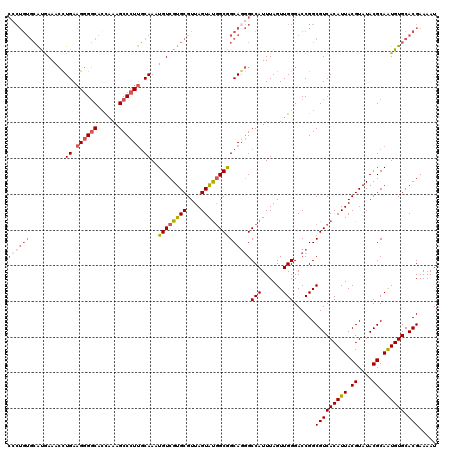
| Location | 21,998,035 – 21,998,155 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.42 |
| Mean single sequence MFE | -39.22 |
| Consensus MFE | -30.24 |
| Energy contribution | -30.68 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21998035 120 - 27905053 AUUUUCGUGCACAUUGCGUAUACGUAAUGUGACGCCGGUCCCAACUAAAUGGCCCUGCUGCCAUACUAACGCACGACAUUUGCAAGGGCUUUGGUGCCCCUUCAGGUUUUAUGCACAGGG ......((((((((((((....))))))))).)))....(((......(((((......)))))......(((..(.(((((...((((......))))...))))).)..)))...))) ( -40.30) >DroSim_CAF1 34359 120 - 1 AUUUUCGUGCACAUUGCGUAUACGUAAUGUGACGCCGGUCCCAACUAAAUGGCCCUGCUGCCAUACUAACGCACGACAUUUGCAAGGGCUUUGGUGCCCCUUCAGGUUUUAUGCACAGGG ......((((((((((((....))))))))).)))....(((......(((((......)))))......(((..(.(((((...((((......))))...))))).)..)))...))) ( -40.30) >DroEre_CAF1 40094 120 - 1 AUUUUCGUGCACAUUGCGUAUACGUAAUGUGACGCCGGUCCCAACUAAAUGGCCAUGCCGCCGUACUAACGCACGACAUUUGCAAGGGCUUUGCUGCCCCUUCAGGCUUCGUGCACAGGG ......((((((((((((....))))))))).)))....(((......(((((......)))))......((((((..((((...((((......))))...))))..))))))...))) ( -44.80) >DroYak_CAF1 38719 120 - 1 AUUUUCGUGCACAUUGCGUAUACGUAAUGUGACGCCGGUCCCAACUAAAUGGCCCUGCCGCCAUACUAACGCACGACAUUUGCAAGGGCUUUGGUGCCCCUUCAGGUUUCAUGCACAGCG ......((.(((((((((....)))))))))))((.(...........(((((......)))))......(((.((.(((((...((((......))))...))))).)).))).).)). ( -37.80) >DroAna_CAF1 44412 112 - 1 AUUUUCGUGCACACUGCGUAUACGUAAUGUGACGCCGGUCCCAACUAAAUGGCUCUGCCGCAACACUAACGCACGACAUUUGGGAUGGCUUCAGUGGCGCAUCAACUUUCAC-------- ......((((.(((((((....))))...(((.((((.((((((......(((...)))((.........)).......)))))))))).))))))))))............-------- ( -32.90) >consensus AUUUUCGUGCACAUUGCGUAUACGUAAUGUGACGCCGGUCCCAACUAAAUGGCCCUGCCGCCAUACUAACGCACGACAUUUGCAAGGGCUUUGGUGCCCCUUCAGGUUUCAUGCACAGGG ......((((((((((((....))))))))).)))((((.(((......)))....))))..........(((.((.(((((...((((......))))...))))).)).)))...... (-30.24 = -30.68 + 0.44)

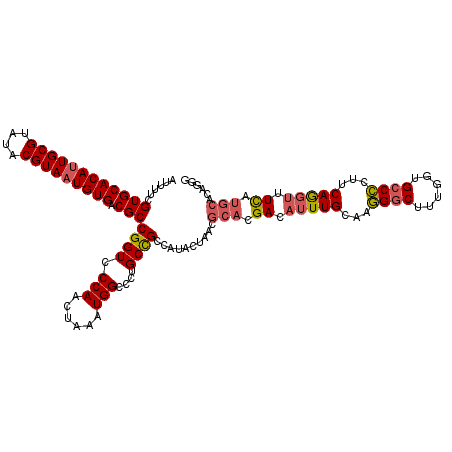
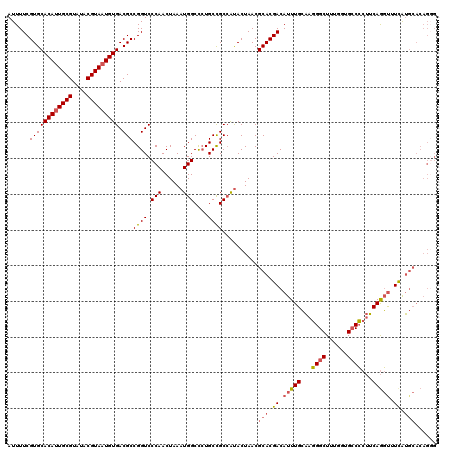
| Location | 21,998,115 – 21,998,234 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.87 |
| Mean single sequence MFE | -41.40 |
| Consensus MFE | -35.31 |
| Energy contribution | -35.79 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21998115 119 - 27905053 GCCCACAAGCACACGUGCAUU-CGCUCGUGUGUUGUAAAAGAGUGGAGUUGGAAUUUGCUUUGCUACGUGACUACUGGUAAUUUUCGUGCACAUUGCGUAUACGUAAUGUGACGCCGGUC ...(((.((((((((.((...-.)).))))))))(((..((((..((.......))..))))..))))))...(((((.......(((.(((((((((....))))))))))))))))). ( -43.31) >DroSim_CAF1 34439 119 - 1 GCCCACAAGCACACGUGCACU-CGCUCGUGUGUAGUAAAAGAGUGGAGUUGGAAUUUGCUUUGCUACGUGACUACUGGUAAUUUUCGUGCACAUUGCGUAUACGUAAUGUGACGCCGGUC ...(((..(((((((.((...-.)).))))))).(((..((((..((.......))..))))..))))))...(((((.......(((.(((((((((....))))))))))))))))). ( -42.91) >DroEre_CAF1 40174 119 - 1 GCCCACAAACACACGUGCACU-UGCUCGUGUGUAGUAAAAGAGUGGAGUUGGAAUUUGCUUUGCUACGUGACUACUGGUAAUUUUCGUGCACAUUGCGUAUACGUAAUGUGACGCCGGUC ...(((..(((((((.((...-.)).))))))).(((..((((..((.......))..))))..))))))...(((((.......(((.(((((((((....))))))))))))))))). ( -42.91) >DroYak_CAF1 38799 119 - 1 ACCCACAAGCACACGUGCACU-CGCUCGUGUGUAGUAAAAGAGUGGAGUUGGAAUUUGCUUUGCUACGUGACUACUGGUAAUUUUCGUGCACAUUGCGUAUACGUAAUGUGACGCCGGUC ...(((..(((((((.((...-.)).))))))).(((..((((..((.......))..))))..))))))...(((((.......(((.(((((((((....))))))))))))))))). ( -42.91) >DroAna_CAF1 44484 120 - 1 CUCCACAUGUGCAAGUACAUACCACACGUUUGUACCGAAAGCGUGGAGUCGGAAUUUGCUUUGGUACGUGACUGUCGGUAAUUUUCGUGCACACUGCGUAUACGUAAUGUGACGCCGGUC ...((((((((((......(((((((.(((((((((.((((((.............)))))))))))).)))))).)))).......)))))..((((....)))))))))......... ( -34.94) >consensus GCCCACAAGCACACGUGCACU_CGCUCGUGUGUAGUAAAAGAGUGGAGUUGGAAUUUGCUUUGCUACGUGACUACUGGUAAUUUUCGUGCACAUUGCGUAUACGUAAUGUGACGCCGGUC ...(((..(((((((.((.....)).))))))).(((..((((..((.......))..))))..))))))...(((((.......(((.(((((((((....))))))))))))))))). (-35.31 = -35.79 + 0.48)

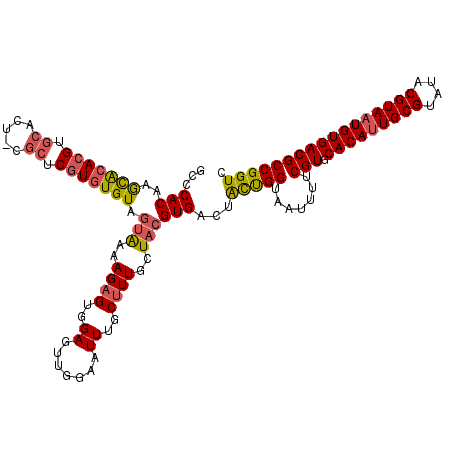
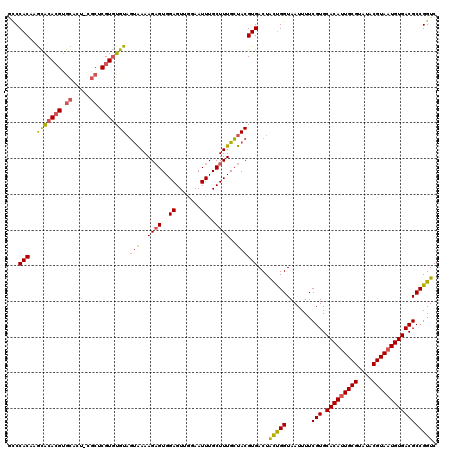
| Location | 21,998,195 – 21,998,312 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.31 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -24.82 |
| Energy contribution | -24.98 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.702612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21998195 117 - 27905053 UGGUCAGACUUGGCAGGAAAAUUGCCCCGAACAAUUAUUAUGAUGGCUGAUAAGCCUCACGCACACU--CUGGCACUCUCGCCCACAAGCACACGUGCAUU-CGCUCGUGUGUUGUAAAA ..((((((...(((((.....)))))..............(((.((((....))))))).......)--)))))..........((((.((((((.((...-.)).)))))))))).... ( -35.70) >DroSim_CAF1 34519 117 - 1 UGGUCAGACUUGGCAGGAAAAUUGCCCCGAACAAUUAUUAUGAUGGCUGAUAAGCCUCACGCACACU--CUGCGACUCUCGCCCACAAGCACACGUGCACU-CGCUCGUGUGUAGUAAAA .(((.(((...(((((.....)))))..............(((.((((....)))))))((((....--.))))..))).))).....(((((((.((...-.)).)))))))....... ( -35.40) >DroEre_CAF1 40254 117 - 1 UGGUCAGACUUGGCAGGAAAAUUGCCCCGAACAAUUAUUAUGAUGGCUGAUAAGCCUCACGCACACU--CAGACACUCUCGCCCACAAACACACGUGCACU-UGCUCGUGUGUAGUAAAA .(((.(((...(((((.....)))))..............(((.((((....)))))))........--.......))).))).....(((((((.((...-.)).)))))))....... ( -31.70) >DroYak_CAF1 38879 119 - 1 UGGUCAGACUUGGCAGGAAAAUUGCCCCGAACAAUUAUUAUGAUGGCUGAUAAGCCUCACGCACACUCGCAGACACUCUCACCCACAAGCACACGUGCACU-CGCUCGUGUGUAGUAAAA .(((.(((...(((((.....)))))..............(((.((((....))))))).((......))......))).))).....(((((((.((...-.)).)))))))....... ( -32.50) >DroAna_CAF1 44564 116 - 1 UGGCUGGACUUGGCAGGAAAAUUGCCCCAAACAAUUAUUAUGAUGGUUGAUAAGCCUCACGCACAUA--CAAACAC--CCCUCCACAUGUGCAAGUACAUACCACACGUUUGUACCGAAA .((((((....(((((.....)))))))...(((((((....)))))))...))))((..((((((.--.......--........))))))..(((((.((.....)).))))).)).. ( -26.69) >consensus UGGUCAGACUUGGCAGGAAAAUUGCCCCGAACAAUUAUUAUGAUGGCUGAUAAGCCUCACGCACACU__CAGACACUCUCGCCCACAAGCACACGUGCACU_CGCUCGUGUGUAGUAAAA .(((.(((...(((((.....)))))..............(((.((((....))))))).................))).))).....(((((((.((.....)).)))))))....... (-24.82 = -24.98 + 0.16)

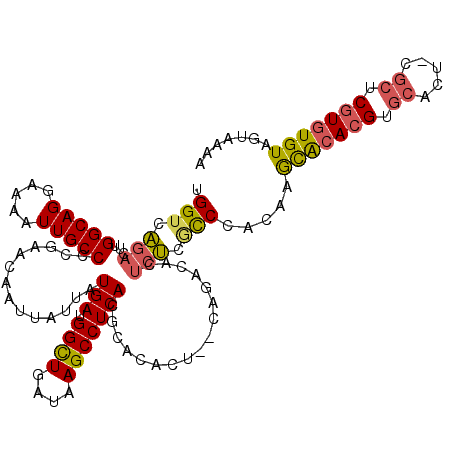
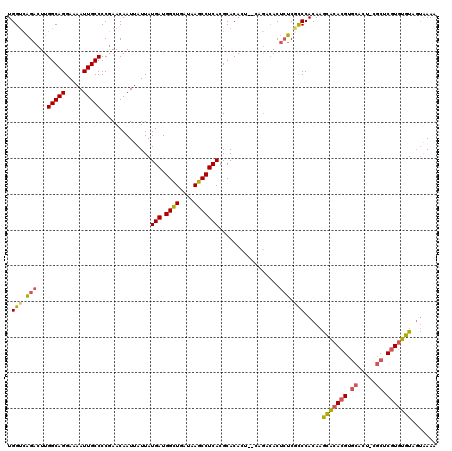
| Location | 21,998,272 – 21,998,391 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.65 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -31.38 |
| Energy contribution | -31.86 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21998272 119 - 27905053 CAACAA-AUUUGGUUUUUAUGACAGCCAAAUGUGUGUGUAAUUAUUCCUGUCGAAUGUCGGAAUGGACGCUGAGGGACCUUGGUCAGACUUGGCAGGAAAAUUGCCCCGAACAAUUAUUA ..(((.-((((((((........)))))))).)))..((((((.((((((((((..(((......)))((..((....))..)).....))))))))))))))))............... ( -34.40) >DroSim_CAF1 34596 119 - 1 CAACAA-AUUUGGUUUUUAUGACAGCCAAAUGUGUGUGUAAUUAUUCCUGUCGAAUGUCGGAAUGGACGCUAAGGGACCUUGGUCAGACUUGGCAGGAAAAUUGCCCCGAACAAUUAUUA ..(((.-((((((((........)))))))).)))..((((((.((((((((((..(((......)))((((((....)))))).....))))))))))))))))............... ( -34.90) >DroEre_CAF1 40331 120 - 1 CAACAAAAUUUCGGUUUUAUGACAGCCAAAUGUGUGUGUAAUUAUUCCUGUCGAAUGUCGGAAUGGACGCUAAGGGACCUUGGUCAGACUUGGCAGGAAAAUUGCCCCGAACAAUUAUUA .........(((((.......(((......)))....((((((.((((((((((..(((......)))((((((....)))))).....)))))))))))))))).)))))......... ( -32.00) >DroYak_CAF1 38958 119 - 1 CAACAA-AUUUGGUUUUUAUGACAGCCAAAUGUGUGUGUAAUUAUUCCUGUCGAAUGUCGGAAUGGACGCUAAGGGACCUUGGUCAGACUUGGCAGGAAAAUUGCCCCGAACAAUUAUUA ..(((.-((((((((........)))))))).)))..((((((.((((((((((..(((......)))((((((....)))))).....))))))))))))))))............... ( -34.90) >DroAna_CAF1 44640 119 - 1 CAACAA-AUUUGGUUUUUAUGACAGCCAAAUGUGUGUGUAAUUAUUCCUGUCGAAUGCCGGAAUGGACGCGAAGGGACCUUGGCUGGACUUGGCAGGAAAAUUGCCCCAAACAAUUAUUA ..(((.-((((((((........)))))))).)))..((((((.((((((((((...((((...((.(.(....)).))....))))..))))))))))))))))............... ( -35.30) >consensus CAACAA_AUUUGGUUUUUAUGACAGCCAAAUGUGUGUGUAAUUAUUCCUGUCGAAUGUCGGAAUGGACGCUAAGGGACCUUGGUCAGACUUGGCAGGAAAAUUGCCCCGAACAAUUAUUA ..(((..((((((((........))))))))...)))((((((.((((((((((..(((......)))((((((....)))))).....))))))))))))))))............... (-31.38 = -31.86 + 0.48)

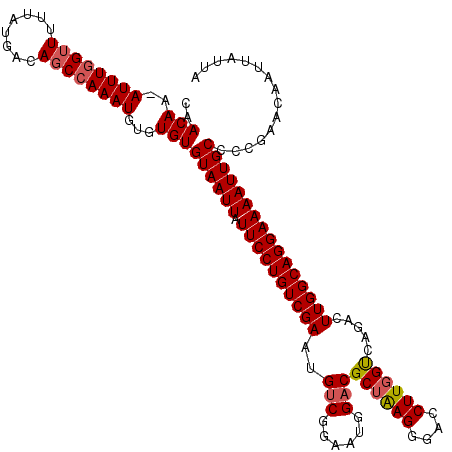
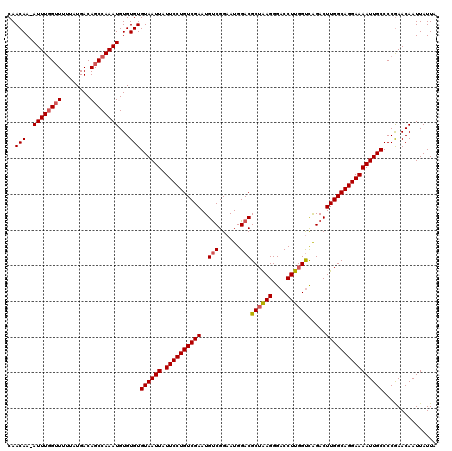
| Location | 21,998,312 – 21,998,430 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.62 |
| Mean single sequence MFE | -22.96 |
| Consensus MFE | -20.61 |
| Energy contribution | -20.65 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21998312 118 + 27905053 AGGUCCCUCAGCGUCCAUUCCGACAUUCGACAGGAAUAAUUACACACACAUUUGGCUGUCAUAAAAACCAAAU-UUGUUGAAAUGU-UCAACUGAAAUGACAGCACAAAGCAUUUACAAC ..........((....(((((...........))))).............((((((((((((.(((......)-))(((((.....-)))))....)))))))).))))))......... ( -22.40) >DroSim_CAF1 34636 118 + 1 AGGUCCCUUAGCGUCCAUUCCGACAUUCGACAGGAAUAAUUACACACACAUUUGGCUGUCAUAAAAACCAAAU-UUGUUGAAAUGU-UCAACUGAAAUGACAGCACAAAGCAUUUACAAC ..........((....(((((...........))))).............((((((((((((.(((......)-))(((((.....-)))))....)))))))).))))))......... ( -22.40) >DroEre_CAF1 40371 119 + 1 AGGUCCCUUAGCGUCCAUUCCGACAUUCGACAGGAAUAAUUACACACACAUUUGGCUGUCAUAAAACCGAAAUUUUGUUGAAAUGU-UCAACUGAAAUGACAGCGCAAAGCAUUUACAAC ..........((....(((((...........))))).............((((((((((((..........(((.(((((.....-))))).))))))))))).))))))......... ( -23.30) >DroYak_CAF1 38998 118 + 1 AGGUCCCUUAGCGUCCAUUCCGACAUUCGACAGGAAUAAUUACACACACAUUUGGCUGUCAUAAAAACCAAAU-UUGUUGAAAUGU-UCAACUGAAAUGACAGCACAAAGCAUUUACAAC ..........((....(((((...........))))).............((((((((((((.(((......)-))(((((.....-)))))....)))))))).))))))......... ( -22.40) >DroAna_CAF1 44680 119 + 1 AGGUCCCUUCGCGUCCAUUCCGGCAUUCGACAGGAAUAAUUACACACACAUUUGGCUGUCAUAAAAACCAAAU-UUGUUGAAAUGUUUCAGCCAAAAUGACAGCACAAAACAUUUACAGC ...(((..(((.((((.....)).)).)))..)))...............((((((((((((..........(-((((((((....))))).)))))))))))).))))........... ( -24.30) >consensus AGGUCCCUUAGCGUCCAUUCCGACAUUCGACAGGAAUAAUUACACACACAUUUGGCUGUCAUAAAAACCAAAU_UUGUUGAAAUGU_UCAACUGAAAUGACAGCACAAAGCAUUUACAAC ..........((....(((((...........))))).............((((((((((((..............(((((......)))))....)))))))).))))))......... (-20.61 = -20.65 + 0.04)

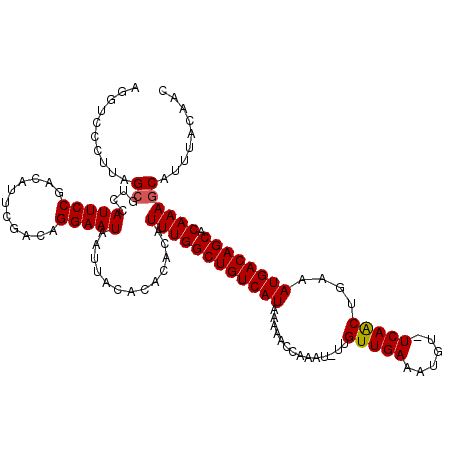
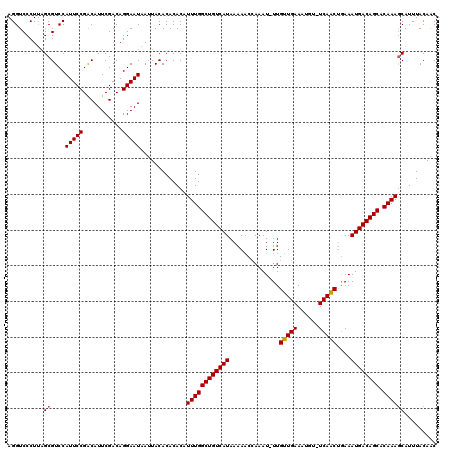
| Location | 21,998,352 – 21,998,460 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -15.71 |
| Energy contribution | -16.15 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21998352 108 + 27905053 UACACACACAUUUGGCUGUCAUAAAAACCAAAU-UUGUUGAAAUGU-UCAACUGAAAUGACAGCACAAAGCAUUUACAAC---------AUUUUGUGUAUUUUGUAUUUGCC-CCCUGCC ..........((((((((((((.(((......)-))(((((.....-)))))....)))))))).))))(((..(((((.---------((.......)).)))))..))).-....... ( -22.70) >DroSim_CAF1 34676 108 + 1 UACACACACAUUUGGCUGUCAUAAAAACCAAAU-UUGUUGAAAUGU-UCAACUGAAAUGACAGCACAAAGCAUUUACAAC---------AUUUUGUGUAUUUUGUAUGUGCC-UCCUGCC ..........((((((((((((.(((......)-))(((((.....-)))))....)))))))).))))((((.(((((.---------((.......)).))))).)))).-....... ( -23.00) >DroEre_CAF1 40411 110 + 1 UACACACACAUUUGGCUGUCAUAAAACCGAAAUUUUGUUGAAAUGU-UCAACUGAAAUGACAGCGCAAAGCAUUUACAAC---------AUUUUGUAUAUUUUGUAUUUGCCCCCCUGCC ..........((((((((((((..........(((.(((((.....-))))).))))))))))).))))(((..(((((.---------((.......)).)))))..)))......... ( -23.60) >DroYak_CAF1 39038 118 + 1 UACACACACAUUUGGCUGUCAUAAAAACCAAAU-UUGUUGAAAUGU-UCAACUGAAAUGACAGCACAAAGCAUUUACAACAUUUUCAACAUUUUGUGUAUUUUGUAUUCGACUCCCUGCC .............((..(((((((((((((((.-.(((((((((((-(......(((((.(........))))))..)))).)))))))).)))).)).))))))....)))..)).... ( -19.70) >DroAna_CAF1 44720 104 + 1 UACACACACAUUUGGCUGUCAUAAAAACCAAAU-UUGUUGAAAUGUUUCAGCCAAAAUGACAGCACAAAACAUUUACAGC---------AUUUUGUUUACUUUCC------CCAUCUACC ..........((((((((((((..........(-((((((((....))))).)))))))))))).))))...........---------................------......... ( -16.80) >consensus UACACACACAUUUGGCUGUCAUAAAAACCAAAU_UUGUUGAAAUGU_UCAACUGAAAUGACAGCACAAAGCAUUUACAAC_________AUUUUGUGUAUUUUGUAUUUGCC_CCCUGCC ((((((....((((((((((((..............(((((......)))))....)))))))).))))(......)................))))))..................... (-15.71 = -16.15 + 0.44)


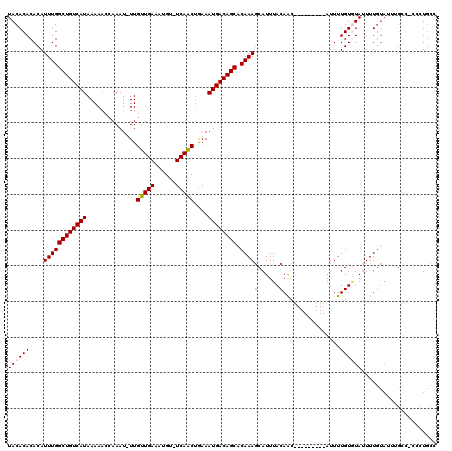
| Location | 21,998,352 – 21,998,460 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.28 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21998352 108 - 27905053 GGCAGGG-GGCAAAUACAAAAUACACAAAAU---------GUUGUAAAUGCUUUGUGCUGUCAUUUCAGUUGA-ACAUUUCAACAA-AUUUGGUUUUUAUGACAGCCAAAUGUGUGUGUA .((....-.)).........(((((((...(---------((((.(((((.((...((((......)))).))-.)))))))))).-((((((((........)))))))).))))))). ( -26.50) >DroSim_CAF1 34676 108 - 1 GGCAGGA-GGCACAUACAAAAUACACAAAAU---------GUUGUAAAUGCUUUGUGCUGUCAUUUCAGUUGA-ACAUUUCAACAA-AUUUGGUUUUUAUGACAGCCAAAUGUGUGUGUA .......-.(((((((((...((((......---------..)))).....((((.((((((((.((((((((-.....)))))..-...))).....))))))))))))))))))))). ( -29.40) >DroEre_CAF1 40411 110 - 1 GGCAGGGGGGCAAAUACAAAAUAUACAAAAU---------GUUGUAAAUGCUUUGCGCUGUCAUUUCAGUUGA-ACAUUUCAACAAAAUUUCGGUUUUAUGACAGCCAAAUGUGUGUGUA .........(((.(((((.....(((((...---------.))))).....((((.((((((((....(((((-.....)))))(((((....)))))))))))))))))))))).))). ( -26.00) >DroYak_CAF1 39038 118 - 1 GGCAGGGAGUCGAAUACAAAAUACACAAAAUGUUGAAAAUGUUGUAAAUGCUUUGUGCUGUCAUUUCAGUUGA-ACAUUUCAACAA-AUUUGGUUUUUAUGACAGCCAAAUGUGUGUGUA (((.....))).........(((((((............(((((.(((((.((...((((......)))).))-.)))))))))).-((((((((........)))))))).))))))). ( -27.20) >DroAna_CAF1 44720 104 - 1 GGUAGAUGG------GGAAAGUAAACAAAAU---------GCUGUAAAUGUUUUGUGCUGUCAUUUUGGCUGAAACAUUUCAACAA-AUUUGGUUUUUAUGACAGCCAAAUGUGUGUGUA .........------..............((---------((((.((((((((((.((((......))))))))))))))))(((.-((((((((........)))))))).))))))). ( -20.00) >consensus GGCAGGG_GGCAAAUACAAAAUACACAAAAU_________GUUGUAAAUGCUUUGUGCUGUCAUUUCAGUUGA_ACAUUUCAACAA_AUUUGGUUUUUAUGACAGCCAAAUGUGUGUGUA ....................(((((((.............((((.(((((......((((......)))).....)))))))))...((((((((........)))))))).))))))). (-19.24 = -19.28 + 0.04)

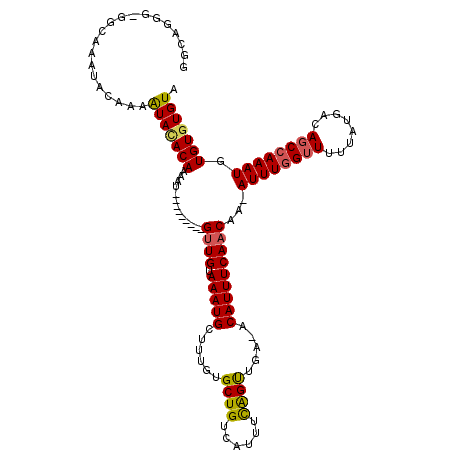
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:44 2006