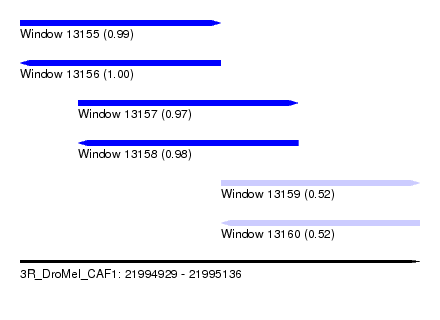
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,994,929 – 21,995,136 |
| Length | 207 |
| Max. P | 0.998616 |

| Location | 21,994,929 – 21,995,033 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.25 |
| Mean single sequence MFE | -24.86 |
| Consensus MFE | -8.97 |
| Energy contribution | -11.17 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.36 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21994929 104 + 27905053 ----ACU----GGGG-GA-AAGCUACCCAGACAAGUUUGAAUGAAAAAUUUCAGCAGCUUCUGUUCUUAUUUUGGAAUGAACA--CAAAA-AAAAAAACAGAAAAUGCUGCUGCCAG ----.((----(((.-..-......)))))..((((((.......))))))((((((((((((((....(((((.........--)))))-.....)))))))...))))))).... ( -27.60) >DroSim_CAF1 31230 107 + 1 ----ACU----GGGG-GA-AAGCUACCCAGACAAGUUUGAAUGAAAAAUUUCAGCAGCUUCUGUUCUUAUUUUGGAAUGAAAAAACAAAAACAAAAAACAGAAAAUGCUGCUGCCAG ----.((----(((.-..-......)))))..((((((.......))))))((((((((((((((.((.(((((...........)))))...)).)))))))...))))))).... ( -26.80) >DroEre_CAF1 36869 109 + 1 ----ACU----GGGGGGAAAAGUCACCCAGACAAGUUUGAAUGAAAAAUUUCAGCAGAUUCUGUUCUUAUUUUGGACUGAAAGGAAAAAAAAAAUAAGAGGAAAGUGCUGCUGCCAG ----.((----(((((((....)).))).......................(((((((((.(.((((((((((...((....))......)))))))))).).))).)))))))))) ( -28.30) >DroYak_CAF1 35510 102 + 1 ----ACA----GGGGAGA-AAGUCACCCAGACAAGUUUGAAUGAAAAAUUUCAGCAGCUUCUGUUCUUAUUUUGGACUGAAA------CAAAAAUAAAAAGAAAAUGCUGCUGCCAG ----...----((...((-((.(((..((((....))))..)))....))))((((((.....(((((.(((((........------))))).....)))))...)))))).)).. ( -22.40) >DroAna_CAF1 41561 105 + 1 CCUUAAUGGUAGCGU-GG-UAGCCA-CCAGACAAGUUAAAAUGAAAAAUUCGAACAGUU--UGCUUUUAGUUUGA--UGCAACAACCA--ACAAAAAAGGGAA---GCAAAAGCCAU .....(((((.(.((-((-...)))-))......(((..(((.....)))..)))..((--(((((((..((((.--((.......))--.))))....))))---))))).))))) ( -19.20) >consensus ____ACU____GGGG_GA_AAGCCACCCAGACAAGUUUGAAUGAAAAAUUUCAGCAGCUUCUGUUCUUAUUUUGGAAUGAAAA_ACAAAAAAAAAAAAAAGAAAAUGCUGCUGCCAG ...........(((...........))).......................(((((((.....(((((.((((....................)))).)))))...))))))).... ( -8.97 = -11.17 + 2.20)


| Location | 21,994,929 – 21,995,033 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.25 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -6.33 |
| Energy contribution | -8.89 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.25 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21994929 104 - 27905053 CUGGCAGCAGCAUUUUCUGUUUUUUU-UUUUG--UGUUCAUUCCAAAAUAAGAACAGAAGCUGCUGAAAUUUUUCAUUCAAACUUGUCUGGGUAGCUU-UC-CCCC----AGU---- (((((((((((...((((((((((..-(((((--.(......)))))).))))))))))))))))).......................(((......-.)-))))----)).---- ( -31.10) >DroSim_CAF1 31230 107 - 1 CUGGCAGCAGCAUUUUCUGUUUUUUGUUUUUGUUUUUUCAUUCCAAAAUAAGAACAGAAGCUGCUGAAAUUUUUCAUUCAAACUUGUCUGGGUAGCUU-UC-CCCC----AGU---- (((((((((((...((((((((((...(((((...........))))).))))))))))))))))).......................(((......-.)-))))----)).---- ( -29.90) >DroEre_CAF1 36869 109 - 1 CUGGCAGCAGCACUUUCCUCUUAUUUUUUUUUUCCUUUCAGUCCAAAAUAAGAACAGAAUCUGCUGAAAUUUUUCAUUCAAACUUGUCUGGGUGACUUUUCCCCCC----AGU---- ((((((((((....(((.(((((((((.................)))))))))...))).)))))).......(((((((........))))))).........))----)).---- ( -22.53) >DroYak_CAF1 35510 102 - 1 CUGGCAGCAGCAUUUUCUUUUUAUUUUUG------UUUCAGUCCAAAAUAAGAACAGAAGCUGCUGAAAUUUUUCAUUCAAACUUGUCUGGGUGACUU-UCUCCCC----UGU---- ..(((((((((.((((.((((((((((.(------.......).)))))))))).))))))))))).......(((((((........)))))))...-.....))----...---- ( -24.90) >DroAna_CAF1 41561 105 - 1 AUGGCUUUUGC---UUCCCUUUUUUGU--UGGUUGUUGCA--UCAAACUAAAAGCA--AACUGUUCGAAUUUUUCAUUUUAACUUGUCUGG-UGGCUA-CC-ACGCUACCAUUAAGG ..(((.(((((---((......((((.--..((....)).--.))))....)))))--))..))).................((((..(((-((((..-..-..))))))).)))). ( -19.90) >consensus CUGGCAGCAGCAUUUUCUGUUUUUUUUUUUUGU_UUUUCAGUCCAAAAUAAGAACAGAAGCUGCUGAAAUUUUUCAUUCAAACUUGUCUGGGUGGCUU_UC_CCCC____AGU____ ....(((((((...((((.(((((.........................))))).))))))))))).......(((((((........)))))))...................... ( -6.33 = -8.89 + 2.56)

| Location | 21,994,959 – 21,995,073 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.60 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -13.80 |
| Energy contribution | -13.88 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21994959 114 + 27905053 AUGAAAAAUUUCAGCAGCUUCUGUUCUUAUUUUGGAAUGAACA--CAAAA-AAAAAAACAGAAAAUGCUGCUGCCAGAAGGCAGCAAUCAGGAAAAGGCGGCACGGACACCACAAGC ........((((((((..(((((((....(((((.........--)))))-.....)))))))..))))((((((....))))))......)))).((.(.......).))...... ( -29.70) >DroSim_CAF1 31260 117 + 1 AUGAAAAAUUUCAGCAGCUUCUGUUCUUAUUUUGGAAUGAAAAAACAAAAACAAAAAACAGAAAAUGCUGCUGCCAGAAGGCAGCAAUCAGGAAAAGGCGGCACGGACACCACAAGC ........((((((((..(((((((.((.(((((...........)))))...)).)))))))..))))((((((....))))))......)))).((.(.......).))...... ( -28.90) >DroEre_CAF1 36901 117 + 1 AUGAAAAAUUUCAGCAGAUUCUGUUCUUAUUUUGGACUGAAAGGAAAAAAAAAAUAAGAGGAAAGUGCUGCUGCCAGAAGGCAGCAAUCAGGACAAGGCGGAACGGACACCACGAGC .((((....))))......((((((((..(((((..((((...................((......))((((((....))))))..))))..))))).)))))))).......... ( -30.10) >DroYak_CAF1 35541 111 + 1 AUGAAAAAUUUCAGCAGCUUCUGUUCUUAUUUUGGACUGAAA------CAAAAAUAAAAAGAAAAUGCUGCUGCCAGAAGGCAGCAAUCAGGACAGGACGGAACGGACACCACGAGC .((((....))))......((((((((..(((((..((((..------...........((......))((((((....))))))..))))..))))).)))))))).......... ( -30.50) >DroAna_CAF1 41598 108 + 1 AUGAAAAAUUCGAACAGUU--UGCUUUUAGUUUGA--UGCAACAACCA--ACAAAAAAGGGAA---GCAAAAGCCAUAAGACAGCAAUCAGGCUCGGGCGACCAGGACAACAGAAGA ........(((......((--(((((((..((((.--((.......))--.))))....))))---))))).(((.......(((......))).((....)).)).)....))).. ( -18.40) >consensus AUGAAAAAUUUCAGCAGCUUCUGUUCUUAUUUUGGAAUGAAAA_ACAAAAAAAAAAAAAAGAAAAUGCUGCUGCCAGAAGGCAGCAAUCAGGAAAAGGCGGCACGGACACCACAAGC ....................((((.(((.((((((........................((......))((((((....))))))..)))))).)))))))................ (-13.80 = -13.88 + 0.08)

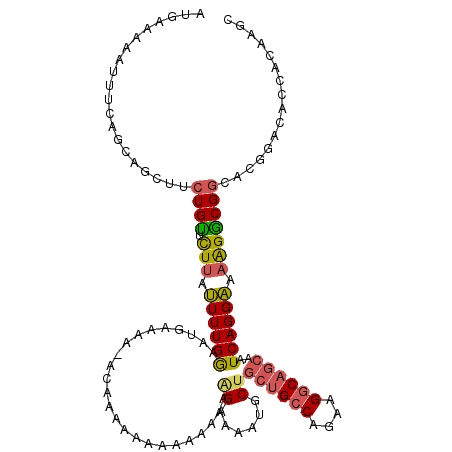
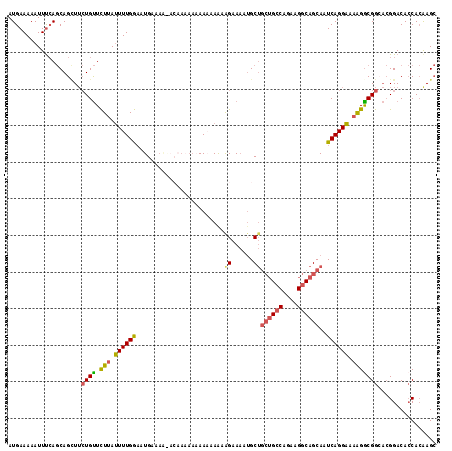
| Location | 21,994,959 – 21,995,073 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.60 |
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -13.71 |
| Energy contribution | -15.83 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21994959 114 - 27905053 GCUUGUGGUGUCCGUGCCGCCUUUUCCUGAUUGCUGCCUUCUGGCAGCAGCAUUUUCUGUUUUUUU-UUUUG--UGUUCAUUCCAAAAUAAGAACAGAAGCUGCUGAAAUUUUUCAU ....(((((......)))))..((((......((((((....))))))((((.(((((((((((..-(((((--.(......)))))).))))))))))).))))))))........ ( -37.00) >DroSim_CAF1 31260 117 - 1 GCUUGUGGUGUCCGUGCCGCCUUUUCCUGAUUGCUGCCUUCUGGCAGCAGCAUUUUCUGUUUUUUGUUUUUGUUUUUUCAUUCCAAAAUAAGAACAGAAGCUGCUGAAAUUUUUCAU ....(((((......)))))..((((......((((((....))))))((((.(((((((((((...(((((...........))))).))))))))))).))))))))........ ( -35.80) >DroEre_CAF1 36901 117 - 1 GCUCGUGGUGUCCGUUCCGCCUUGUCCUGAUUGCUGCCUUCUGGCAGCAGCACUUUCCUCUUAUUUUUUUUUUCCUUUCAGUCCAAAAUAAGAACAGAAUCUGCUGAAAUUUUUCAU ....((((........)))).......(((..((((((....))))))((((..(((.(((((((((.................)))))))))...)))..))))........))). ( -27.13) >DroYak_CAF1 35541 111 - 1 GCUCGUGGUGUCCGUUCCGUCCUGUCCUGAUUGCUGCCUUCUGGCAGCAGCAUUUUCUUUUUAUUUUUG------UUUCAGUCCAAAAUAAGAACAGAAGCUGCUGAAAUUUUUCAU ....(..((.((.((((.....((..((((.(((((((....)))))))(((...............))------).))))..))......)))).)).))..)((((....)))). ( -28.06) >DroAna_CAF1 41598 108 - 1 UCUUCUGUUGUCCUGGUCGCCCGAGCCUGAUUGCUGUCUUAUGGCUUUUGC---UUCCCUUUUUUGU--UGGUUGUUGCA--UCAAACUAAAAGCA--AACUGUUCGAAUUUUUCAU .......(((..(.((..((..(((((.(((....)))....)))))..))---..))....(((((--(....(((...--...)))....))))--))..)..)))......... ( -17.80) >consensus GCUUGUGGUGUCCGUGCCGCCUUGUCCUGAUUGCUGCCUUCUGGCAGCAGCAUUUUCUGUUUUUUUUUUUUGU_UUUUCAGUCCAAAAUAAGAACAGAAGCUGCUGAAAUUUUUCAU ....((((........)))).......(((..((((((....))))))((((.(((((.(((((.........................))))).))))).))))........))). (-13.71 = -15.83 + 2.12)

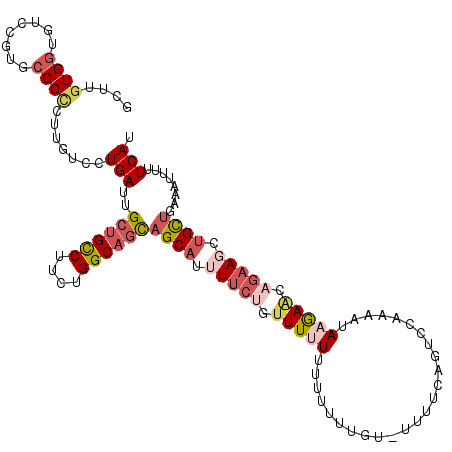
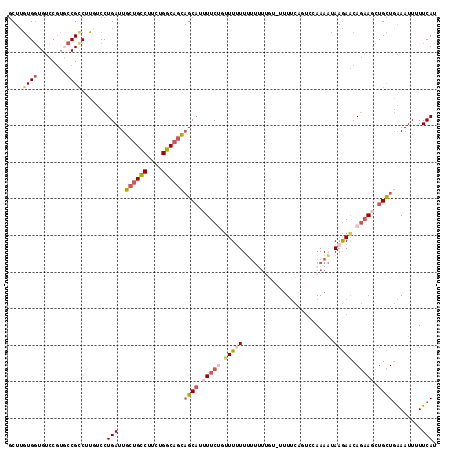
| Location | 21,995,033 – 21,995,136 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.91 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -16.41 |
| Energy contribution | -16.01 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21995033 103 + 27905053 AAGGCAGCAAUCAGGAAAAGGCGGCACGGACACCACAAGCAACUAAAAAUGUCCUGCCACAUUUUCG---------------CACACA--GGGAAAAGUGAGCUUUUUUGCGGCUUAAAA .((((.((((....(((((.(.((((.(((((.................))))))))).).)))))(---------------(.(((.--.......))).))....)))).)))).... ( -29.23) >DroSim_CAF1 31337 103 + 1 AAGGCAGCAAUCAGGAAAAGGCGGCACGGACACCACAAGCAACUAAAAAUGUCCUGCCACAUUUUCG---------------CACACA--GGGAAAAGUGAGCUUUUUUGCGGCUUAAAA .((((.((((....(((((.(.((((.(((((.................))))))))).).)))))(---------------(.(((.--.......))).))....)))).)))).... ( -29.23) >DroEre_CAF1 36978 102 + 1 AAGGCAGCAAUCAGGACAAGGCGGAACGGACACCACGAGCAACUAAAAAUGUCCUGCCACAUUUUCG---------------CACACA--GGGAAAAGUGAGCUU-UUUGCGGCUUAGAA .((((.((((..((((((.((..(..((.......))..)..)).....))))))(((((.(((((.---------------(.....--).)))))))).))..-.)))).)))).... ( -26.50) >DroYak_CAF1 35612 102 + 1 AAGGCAGCAAUCAGGACAGGACGGAACGGACACCACGAGCAACUAAAAAUGUCCUGCCACAUUUUCG---------------CACACA--GGGAAAAGUGAGCUU-UUUGCGGCUUAAAA .((((.((((..((((((.....(..((.......))..).........))))))(((((.(((((.---------------(.....--).)))))))).))..-.)))).)))).... ( -25.74) >DroAna_CAF1 41666 119 + 1 AAGACAGCAAUCAGGCUCGGGCGACCAGGACAACAGAAGAAACUUAAAAUGUCCUGCCCCUUAUAUGUGCCUGUGUGGUCGGCAGACUUUGGAGCUGGCCAACUU-GUUGUGGUUUAAAA ...(((((((.(((((.(((((....((((((....(((...)))....)))))))))).......).)))))..((((((((..........))))))))..))-)))))......... ( -38.41) >consensus AAGGCAGCAAUCAGGAAAAGGCGGCACGGACACCACAAGCAACUAAAAAUGUCCUGCCACAUUUUCG_______________CACACA__GGGAAAAGUGAGCUU_UUUGCGGCUUAAAA .((((.((((..(((....((((....(((((.................)))))))))..........................(((..........)))..)))..)))).)))).... (-16.41 = -16.01 + -0.40)

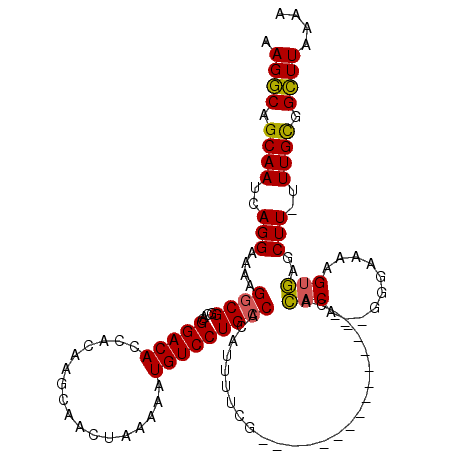
| Location | 21,995,033 – 21,995,136 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.91 |
| Mean single sequence MFE | -31.46 |
| Consensus MFE | -17.38 |
| Energy contribution | -17.46 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.515392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21995033 103 - 27905053 UUUUAAGCCGCAAAAAAGCUCACUUUUCCC--UGUGUG---------------CGAAAAUGUGGCAGGACAUUUUUAGUUGCUUGUGGUGUCCGUGCCGCCUUUUCCUGAUUGCUGCCUU ......((.((((....((.(((.......--.))).)---------------)(((((.(((((((((((((...((...))...))))))).)))))).)))))....)))).))... ( -35.10) >DroSim_CAF1 31337 103 - 1 UUUUAAGCCGCAAAAAAGCUCACUUUUCCC--UGUGUG---------------CGAAAAUGUGGCAGGACAUUUUUAGUUGCUUGUGGUGUCCGUGCCGCCUUUUCCUGAUUGCUGCCUU ......((.((((....((.(((.......--.))).)---------------)(((((.(((((((((((((...((...))...))))))).)))))).)))))....)))).))... ( -35.10) >DroEre_CAF1 36978 102 - 1 UUCUAAGCCGCAAA-AAGCUCACUUUUCCC--UGUGUG---------------CGAAAAUGUGGCAGGACAUUUUUAGUUGCUCGUGGUGUCCGUUCCGCCUUGUCCUGAUUGCUGCCUU ......((((((..-..((.(((.......--.))).)---------------).....))))))((((((......(.....)((((........))))..))))))............ ( -26.80) >DroYak_CAF1 35612 102 - 1 UUUUAAGCCGCAAA-AAGCUCACUUUUCCC--UGUGUG---------------CGAAAAUGUGGCAGGACAUUUUUAGUUGCUCGUGGUGUCCGUUCCGUCCUGUCCUGAUUGCUGCCUU ......((.((((.-..((.(((.......--.))).)---------------)........((((((((.......(.....)..((........))))))))))....)))).))... ( -25.70) >DroAna_CAF1 41666 119 - 1 UUUUAAACCACAAC-AAGUUGGCCAGCUCCAAAGUCUGCCGACCACACAGGCACAUAUAAGGGGCAGGACAUUUUAAGUUUCUUCUGUUGUCCUGGUCGCCCGAGCCUGAUUGCUGUCUU .........(((.(-((((((((..(((....)))..))))))....(((((.(......)((((((((((.....((......))..))))))....))))..))))).))).)))... ( -34.60) >consensus UUUUAAGCCGCAAA_AAGCUCACUUUUCCC__UGUGUG_______________CGAAAAUGUGGCAGGACAUUUUUAGUUGCUUGUGGUGUCCGUGCCGCCUUGUCCUGAUUGCUGCCUU ......((.((((.......(((..........)))........................(((((((((((((.............))))))).))))))..........)))).))... (-17.38 = -17.46 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:33 2006