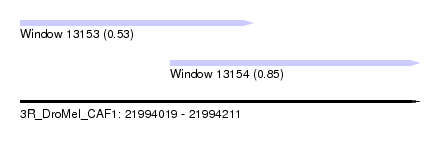
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 21,994,019 – 21,994,211 |
| Length | 192 |
| Max. P | 0.847642 |

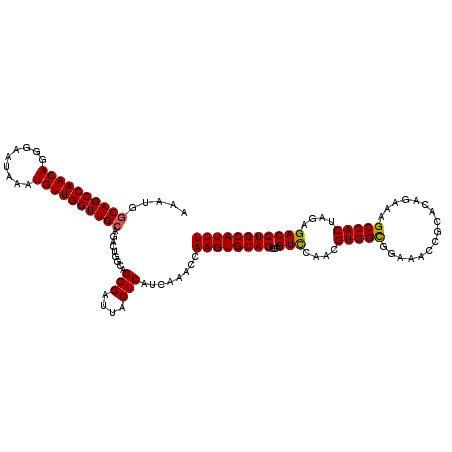
| Location | 21,994,019 – 21,994,131 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.79 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -22.75 |
| Energy contribution | -23.12 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21994019 112 + 27905053 AAAU--------AAAUAUAUGUCAGCGGGCAUUAUGCUUUACGGUUUUCUCAUUGCUGCAGACCUGGCCUCGUCCUCGGAAAAUGGCAGCCAACAGGGAAUAAAUGUUGGUUGCAGACUG ....--------........(((.(.((((.....(((....(((((.(........).))))).)))...)))).)........((((((((((.........)))))))))).))).. ( -29.50) >DroSim_CAF1 30308 111 + 1 AAA---------AAAUAUAUGUCAGCGGGCAUUAUCCUUUACGGUUUUCUCAUUGCGGAAGACCUGGCCUCGUCCGCGGAAAAUGGCAGCCAACAGGGAAUAAAUGUUGGUUGCAGACUG ...---------........(((.((((((.....((.....((((((((......)))))))).))....))))))........((((((((((.........)))))))))).))).. ( -38.30) >DroEre_CAF1 35970 112 + 1 AACA--------AAAUAUAUGUCAGGGGGCAUUAUACUUUAAUGUUUUCUCAUUGCGCCAGACUUGGCUUCGUCCGCAGAAAAUGGCAGCCAACAGGGAAUAAAUGUUGGUUGCAGACUG ....--------........(((((..(((((((.....)))))))..))..(((((...(((........))))))))......((((((((((.........)))))))))).))).. ( -34.00) >DroYak_CAF1 34596 120 + 1 AAAAAAUAAGCGAAAUAUAUGUCAGCAGCCAUUAUACUUUAAUGUUUUCUCAUUGCGGCAGACUUGGCCACGUCCGUGGAAAAUGACAGCCAACAGGGAAUAAAUGUUGGUUGCAGACUG .........(((.......(((((....((((...((..(((((......))))).(((.......)))..))..))))....)))))(((((((.........))))))))))...... ( -26.70) >consensus AAAA________AAAUAUAUGUCAGCGGGCAUUAUACUUUAAGGUUUUCUCAUUGCGGCAGACCUGGCCUCGUCCGCGGAAAAUGGCAGCCAACAGGGAAUAAAUGUUGGUUGCAGACUG ....................(((....................(((((((....(((...(((........))))))))))))).((((((((((.........)))))))))).))).. (-22.75 = -23.12 + 0.37)

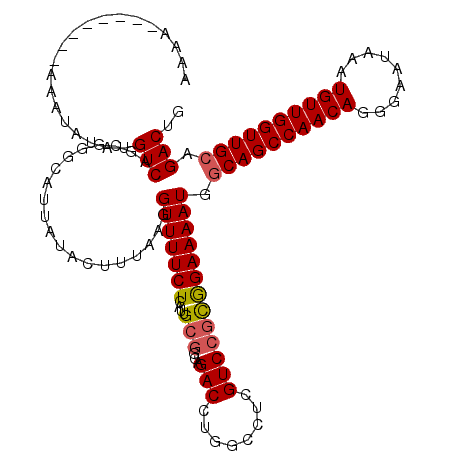
| Location | 21,994,091 – 21,994,211 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -31.99 |
| Energy contribution | -31.86 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 21994091 120 + 27905053 AAAUGGCAGCCAACAGGGAAUAAAUGUUGGUUGCAGACUGGCAGGGAUUACUCAUCAAACCAAGUGCAGUCAGUUCAACGUGGCGGAAACCGCACAUAAAGCCACUAGAGACCUGCAUUU .....((((((((((.........))))))))))......((((((((.....))).......((((.((((........))))(....).))))................))))).... ( -35.60) >DroSim_CAF1 30379 120 + 1 AAAUGGCAGCCAACAGGGAAUAAAUGUUGGUUGCAGACUGUCAGGGAUUACUCAUCAAACCAAGUGCAGGCAGUCCAACGUGGCGGAAACCGCACAGAAAGCCACUAGAGACCUGCAUUU .....((((((((((.........)))))))))).........(((....)))........((((((((((.(.....)((((((....)(.....)...)))))....).))))))))) ( -36.10) >DroEre_CAF1 36042 120 + 1 AAAUGGCAGCCAACAGGGAAUAAAUGUUGGUUGCAGACUGGCAGGGAUUACUCAUCAAACCAAGUGCAGUCUGUCCAACGUGGUGGAAACCGCACAGAAAGCCACUAGAGACCUGCAUUU .....((((((((((.........))))))))))......((((((((.....)))......(((((..(((((.....(((((....))))))))))..).)))).....))))).... ( -41.80) >DroYak_CAF1 34676 120 + 1 AAAUGACAGCCAACAGGGAAUAAAUGUUGGUUGCAGACUGGCAGGGAUUACUCAUCAAACCAAGUGCAGUAAGUCCAACGUGGCGGAAACCGCACAGAAAGCCACUAGAGACCUGCAUUU ...((.(((((((((.........))))))))))).....(((((((((((((((........))).))).)))))...((((((....)(.....)...))))).......)))).... ( -35.70) >consensus AAAUGGCAGCCAACAGGGAAUAAAUGUUGGUUGCAGACUGGCAGGGAUUACUCAUCAAACCAAGUGCAGUCAGUCCAACGUGGCGGAAACCGCACAGAAAGCCACUAGAGACCUGCAUUU .....((((((((((.........)))))))))).........(((....)))........((((((((...(((....(((((................)))))....))))))))))) (-31.99 = -31.86 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:04:28 2006