
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 2,794,735 – 2,794,826 |
| Length | 91 |
| Max. P | 0.829640 |

| Location | 2,794,735 – 2,794,826 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.67 |
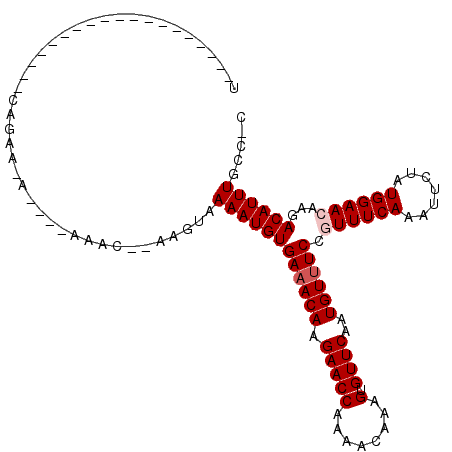
| Mean single sequence MFE | -17.53 |
| Consensus MFE | -13.20 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 2794735 91 + 27905053 UG------------------CCAAGGA----AAAC--AAGUAAAAUGUGAAACAAGAACCAAAACAAAGUGUUCGAUGUUUCCGUUUCAAAUUCUGUGGAAGAAGACAUUUGCC-G ..------------------....((.----....--.....(((((.((((((.(((((........).))))..)))))))))))((((((((........))).)))))))-. ( -16.40) >DroVir_CAF1 122034 107 + 1 UUUAGCAACAGCGGCAGCAGCAGU--A----AAAA--AAGCAAAAUGUGAUACAAGAACCAAAACAAAGUGUUCAAUGUUUCCGUUUCAAAUUCUAUGGAACAAGACAUUUGUG-C ....((....)).((....))...--.----....--..(((((((((((.(((.(((((........).))))..))).)).((((((.......))))))...)))))).))-) ( -16.30) >DroGri_CAF1 118185 97 + 1 UUCAGC------------AGCAGAAAA----AAAG--AGGUAAAAUGUGAAACAAGAACCAAAACAAAGUGUUCAAUGUUUCCGUUUCAAAUUCUAUGGAACAAGACAUUUGCG-C ....((------------.(((((...----..((--((...(((((.((((((.(((((........).))))..)))))))))))....)))).((........))))))))-) ( -17.80) >DroWil_CAF1 89578 92 + 1 U--------------------UGAAUA----AAACGAAGGGAAAAUGUGAAACAAGAACCAAAACAAAGUGUUCAAUGUUUCCGUUUCAAAUUCUAUGGAACAAGACAUUUGCCCG .--------------------......----.......(((.((((((((((((.(((((........).))))..)))))).((((((.......))))))...)))))).))). ( -22.00) >DroMoj_CAF1 126289 83 + 1 --------------------------A----AAAA--AAGGAAAAUGUGAAACAAGAACCAAAACAAAGUGUUCAAUGUUUCCGUUUCAAAUUCUAUGGAACAAGACAUUUGCG-C --------------------------.----....--...(.((((((((((((.(((((........).))))..)))))).((((((.......))))))...)))))).).-. ( -15.20) >DroAna_CAF1 65741 95 + 1 UG------------------CAAAAAAAAAGAAAC--AAGUAAAAUGUGAAACAAGAACCAAAACAAAGUGUUCGAUGUUUCCGUUUCAAAUUCUGUGGAAGAAGACAUUUGCC-G .(------------------((((...........--.....(((((.((((((.(((((........).))))..)))))))))))....((((.....))))....))))).-. ( -17.50) >consensus U___________________CAGAA_A____AAAC__AAGUAAAAUGUGAAACAAGAACCAAAACAAAGUGUUCAAUGUUUCCGUUUCAAAUUCUAUGGAACAAGACAUUUGCC_C ..........................................((((((((((((.(((((........).))))..)))))).((((((.......))))))...))))))..... (-13.20 = -13.70 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:54 2006